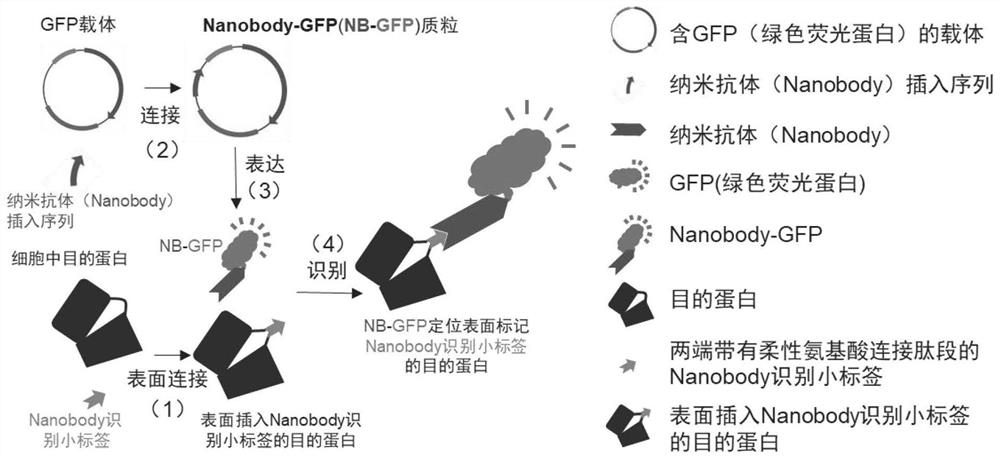
Target protein labeling or tracing composition and method
A target protein and composition technology, which is applied in the field of target protein labeling or tracer composition, can solve problems such as affecting the function of the target protein, lack of function of the target protein, affecting the structural stability of the target protein, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1 Dnm1 tracer
[0078] 1.1 The N- or C-terminal fluorescent labeling of Dnm1 affects its function
[0079] In fission yeast, Dnm1 is the only protein known to directly mediate mitochondrial fission. Our study found that the mitochondrial morphology of wild-type yeast usually forms a complex network structure, and the number of mitochondria in each cell is generally multiple. When Dnm1 is deleted in fission yeast, mitochondria do not divide, and the mitochondria are abnormally morphologically and number 1 (see figure 2 A); The statistical graph of the number of mitochondria in each cell is figure 2 B. When the C- and N-termini of Dnm1 were tagged with GFP or HA, the mitochondrial phenotype was identical to that in the absence of Dnm1 (see figure 2 A. figure 2 B). When the GFP tag was replaced with two red fluorescent protein tags, mCherry or tdTomato, the mitochondrial phenotype was also the same as that in the absence of Dnm1 ( figure 2 B). It can b...
Embodiment 2
[0086] Example 2 Mal3 tracer
[0087] Mal3 is a microtubule positive end-binding protein in fission yeast and maintains microtubule stability.
[0088] 2.1 The N- or C-terminal fluorescent labeling of Mal3 affects its function
[0089] We found that C- or N-terminal labeling of Mal3 resulted in very short microtubules, similar to the microtubule phenotype of Mal3-deficient cells (see Figure 5 A, B). Thus, Mal3 C-terminal or N-terminal tagged GFP can affect Mal3 function. When Mal3 is out of function or functionally impaired, microtubules are unstable and prone to premature depolymerization, making it difficult to grow to wild-type microtubule lengths, thereby exhibiting a phenotype with shorter microtubules.
[0090] 2.2 Design of Mal3 surface fluorescent labeling
[0091] Since the labeling of both ends of Mal3 affects protein function, we tried to label Mal3 with a new method. First, we obtained the resolved structure of Schizosaccharomyces cerevisiae Mal3 (PDB: 5m79) ...
Embodiment 3
[0094] Example 3 Nda3 localization observation
[0095] Microtubules are formed by the polymerization of alpha microtubules and beta tubulin. In Schizosaccharomyces cerevisiae, α-tubulin Nda2 and β-tubulin Nda3 are essential genes, and the two ends of the proteins are labeled and lethal. Therefore, most researchers can only label the non-essential gene at the N-terminus of α-tubulin Atb2 (the C-terminus cannot also be labeled) to achieve microtubule observation. We used the novel method invented here to label the beta-tubulin Nda3 and assess changes in microtubule function.
[0096] 3.1 Design of a new method to label β-tubulin
[0097] First, we obtained the resolved fission yeast Nda3 complex structure (PDB: 5mjs) according to the website https: / / www.rcsb.org / ; according to the Nda3 structure (see Figure 8 ), we searched for the loop sequence facing outward on the surface of the target protein at the interface where the target protein does not interact with its interacti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com