Molecular marker SSR primer for identifying genetic relationship and variety of rehmannia glutinosa variety, kit and application
A genetic relationship and molecular marker technology, which is applied in the field of molecular marker SSR primers for species identification, and the genetic relationship of Rehmannia glutinosa varieties, can solve the problems of inability to construct variety fingerprints, failure to achieve specific identification of a single variety, etc. Increased stability and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
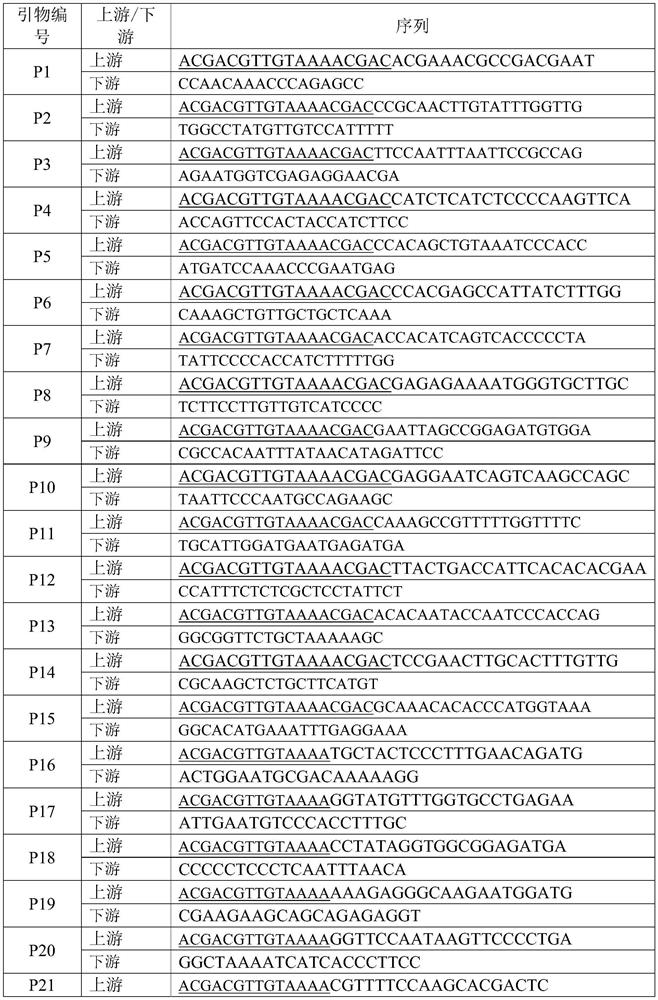
[0028] This example provides 50 pairs of SSR primers, whose sequences are as shown in SEQ ID NO. 1-100.
Embodiment 2
[0029] Example 2 PCR reaction
[0030] 2.1 Sample collection and DNA extraction
[0031]For each of the above 17 cultivars, 3 Rehmannia glutinosa plants with the same growth and size were selected, and 3 to 5 pieces of young leaves from each plant were temporarily stored on dry ice. Total DNA was extracted by CTAB method, and DNA was determined by 0.8% agarose electrophoresis and microspectrophotometer. All samples were diluted to 50ng / μL and stored at -20°C for later use;
[0032] 2.2 Preparation of synthetic TP-M13-SSR primers
[0033] Synthesize the 50 pairs of SSR primer sequences provided in Example 1 above, and connect the 5 ends of the upstream primers of all primer pairs to the M13 sequence (ACACGACGTTGTAAAACGAC) to form 50 pairs of TP-M13 primers. The primers were diluted to 1pm / μL (upstream) and 10pm / μL (downstream) for use; FAM, PET, VIC and NED fluorophores were introduced into the M13 sequence to provide four fluorescent primers, which were diluted to 10pm / μL fo...
Embodiment 3
[0041] Example 3 PCR product analysis and data processing
[0042] 3.1 Electrophoresis detection
[0043] Firstly, the PCR products corresponding to each variety obtained in Example 2 were subjected to electrophoresis detection. The specific steps included: after the PCR products labeled with FAM, PET, VIC and NED fluorescence were evenly mixed in a ratio of 2:4:2:2, from Pipet 1 μL of the mixture and add it to the wells of the deep-well plate dedicated to the DNA analyzer. Add 0.1 μL of LIZ500 molecular weight internal standard and 8.9 μL of deionized formamide to each well of the plate; denature the samples at 95 °C for 5 min on the PCR instrument, and take them out. , immediately placed on crushed ice, cooled for more than 10 min, centrifuged briefly for 10 s, and placed on a DNA analyzer (ABI3730);
[0044] 3.2 Analysis of results
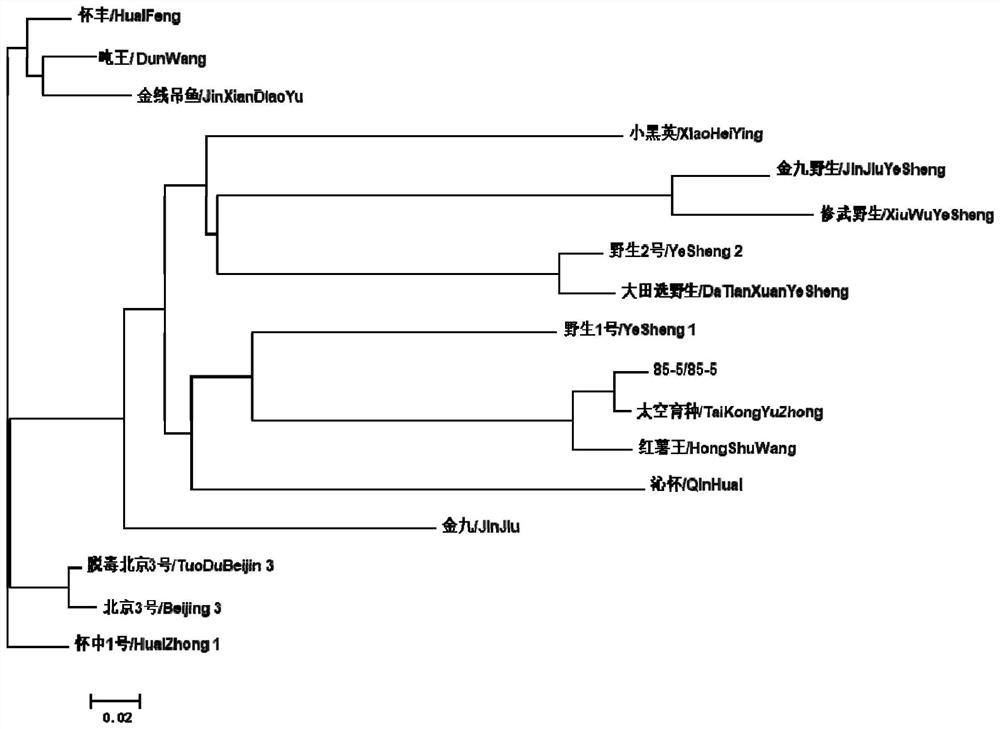
[0045] 3.2.1 Cluster analysis among varieties
[0046] After summarizing the amplified fragments of 50 SSR primers, the software TASSEL3.0 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com