Drug-target interaction prediction algorithm based on graph convolution and similarity
A prediction algorithm and similarity technology, applied in the fields of chemical property prediction, genomics, biological systems, etc., can solve the problems of unpredictable drug-target interactions and complex biological systems, so as to accelerate the process of drug development and improve the overall Efficiency, the effect of reducing feature loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The following technical solutions of the present invention are further elaborated in conjunction with specific embodiments.
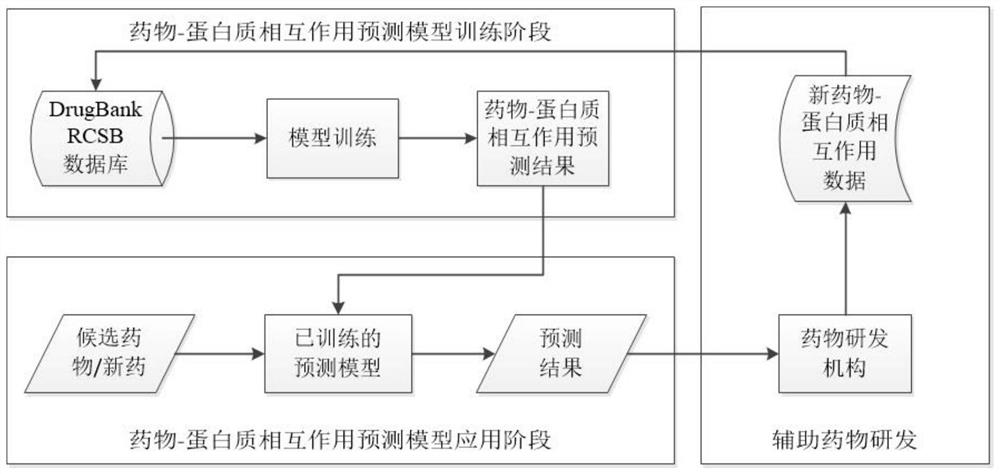
[0029] The theme scheme of this system mainly embodies the basic idea of feature extraction method based on graph convolution and similarity, so as to rapidly predict drug-target interactions to assist in accelerating drug development. A drug-target interaction prediction algorithm based on graph convolution and similarity, including intermolecular structure information extraction module, molecular graph structure information extraction module and drug-target interaction prediction module, the basic steps are as follows:
[0030] 1) Get DTI data from the DugBank database. Each sample contains a drug protein pair and an annotated DTIs. The drug was identified again in The DrugBank to collect its SMILES representation. Proteins are identified in the RCSB protein database, collecting their standard structure PDB files;
[0031] 2) The intermolecular struc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com