Label-free assessment of biomarker expression using vibration spectroscopy
A biomarker, vibrational spectroscopy technique for label-free assessment of biomarker expression using vibrational spectroscopy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
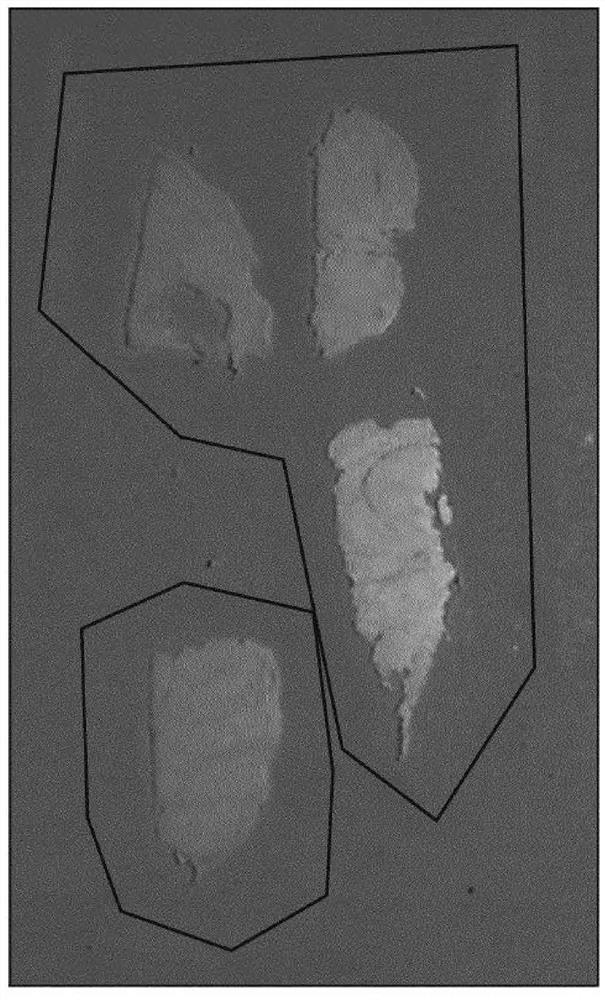
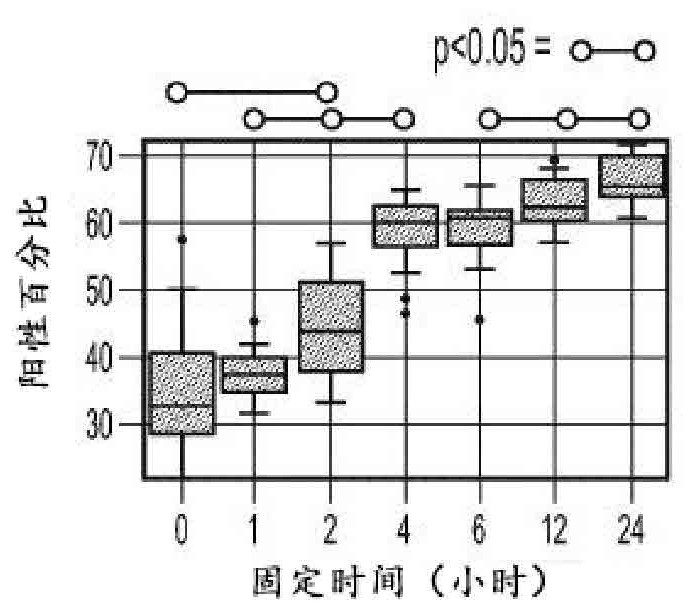
[0198] The expression and fixation time of three different biomarkers (BCL2, FOXP3, and ki67) are presented here. Stain each fixed time tissue block for each biomarker and quantify expression across the slide using an image analysis algorithm (e.g., an algorithm suitable for quantitatively determining the expression level of each stain, For example, an automated algorithm that first segments the tissue on a slide and then identifies regions of the tissue that are not of interest; the algorithm will then automatically determine whether the tissue is positive or negative for a given protein biomarker). Figures 9A, 9B, and 9C show summary results for BCL2, ki-67, and FOXP3, respectively, in the form of box-and-whisker plots and at fixed times. BCL2 and FOXP3 were found to be particularly unstable and susceptible to inappropriate fixation, with their expression levels increasing steadily and monotonically with fixation time.
[0199] On the other hand, ki-67 was found to be relat...
example 2
[0202] MirrIR microscope slides (Kevley Technologies, Chesterland, OH) for reflectance infrared studies were used for mid-IR spectroscopy measurements. Four-micron serial sections of formalin-fixed, paraffin-embedded (FFPE) tonsil tissue were mounted on pretreated MirrIR slides. Dewax the tonsil tissue manually according to OP2100-025. Briefly, after the xylene step, slides were hydrated through a decreasing gradient of ethanol and then transferred to a Rapid Antigen Retrieval (RAR) bench in VENTANA Cell Conditioner 1 (CC1) solution.
[0203] Perform antigen retrieval in CC1 solution in the RAR chamber, which is prepressurized to 30 psi before turning on the heater. The total heating time for any given experiment consists of a 90 s ramp up time and a 2 min cool down time. After the antigen retrieval step, slides were gently washed in deionized water and air dried at room temperature. Dried slides with intact tonsil tissue were used for mid-IR measurements. Single antigen r...
example 3
[0207] Example 3 – Estimating Biomarker Expression Using a Trained Biomarker Expression Estimation Engine
[0208] the summary of instruction
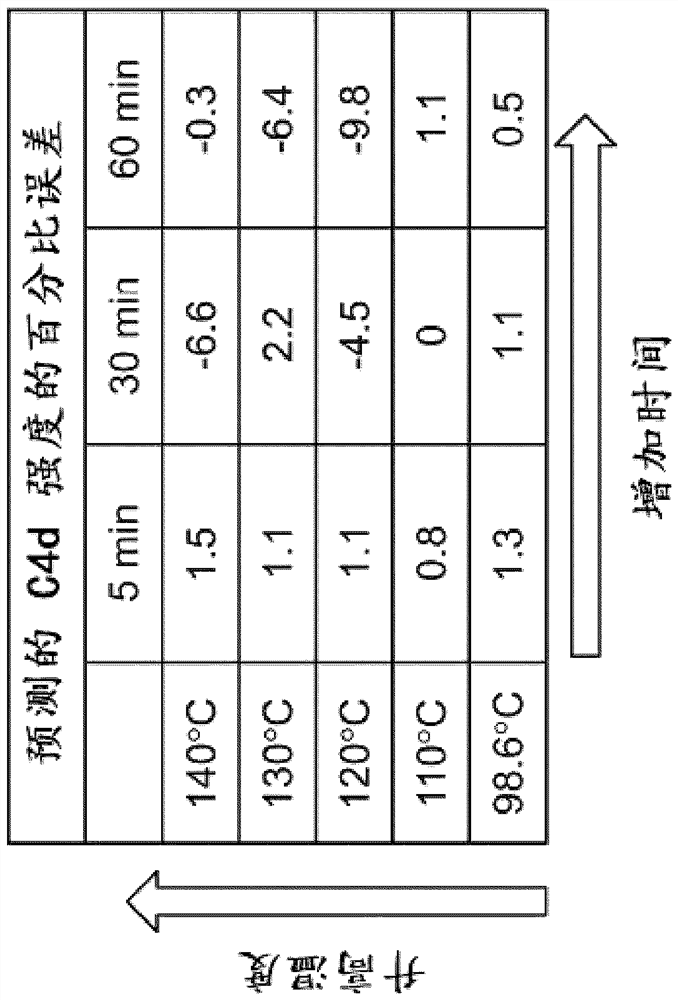
[0209] This experiment utilizes mid-infrared (mid-IR) spectroscopy to examine the vibrational state of molecules in histological tissue sections. In this work, changes in mid-IR spectra caused by differential repair of tonsil tissue are investigated and used to train a biomarker expression estimation engine. Identified shifts in the Mid-IR spectrum correlate with immunohistochemical (IHC) staining for Ki-67 and C4d proteins.
[0210] introduce
[0211] Mid-infrared spectroscopy (mid-IR) is a powerful optical technique that probes the vibrational states of individual molecules in tissues and is very sensitive to the conformational state of proteins. This extreme sensitivity makes mid-IR spectroscopy ideal for microscopy applications, as the presence of endogenous and exogenous materials and even conformational states can be rev...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com