Preparation method for inducing bacteria to form small colony variants
A technology of small colonies and variants, applied in the field of microorganisms, can solve the problems of easy coverage by wild strains, large workload of SCVs, and difficulty in separation and purification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] The K-B disc method provided by the invention induces the test method of Escherichia coli to form SCVs, uses following reagent:
[0038] Sterile filter paper discs containing 30 μg, 60 μg, 120 μg and 240 μg of kanamycin, respectively;
[0039] Sterile filter paper discs containing 5 μg, 10 μg, 20 μg and 40 μg of gentamycin respectively;
[0040] Trypsin Soy Agar (TSA), Trypsin Soy Broth (TSB) and Columbia Blood Agar (referred to as blood plate), Nutrient Agar (NA).
[0041]The main components of trypticase soybean agar medium: casein trypsin hydrolyzate 15.0g / L, soybean powder papain hydrolyzate 5.0g / L, sodium chloride 5.0g / L, agar 15.0g / L, pH value 7.3±0.2 .
[0042] The main components of trypticase soybean broth medium: casein trypsin hydrolyzate 15.0g / L, soybean powder papain hydrolyzate 5.0g / L, sodium chloride 5.0g / L, pH value 7.3±0.2.
[0043] Main components of Columbia blood agar medium: Add 5-10% sterile sheep whole blood on the basis of TSA.
[0044] Main ...
Embodiment 2
[0056] Specific gene detection of Escherichia coli and its SCVs
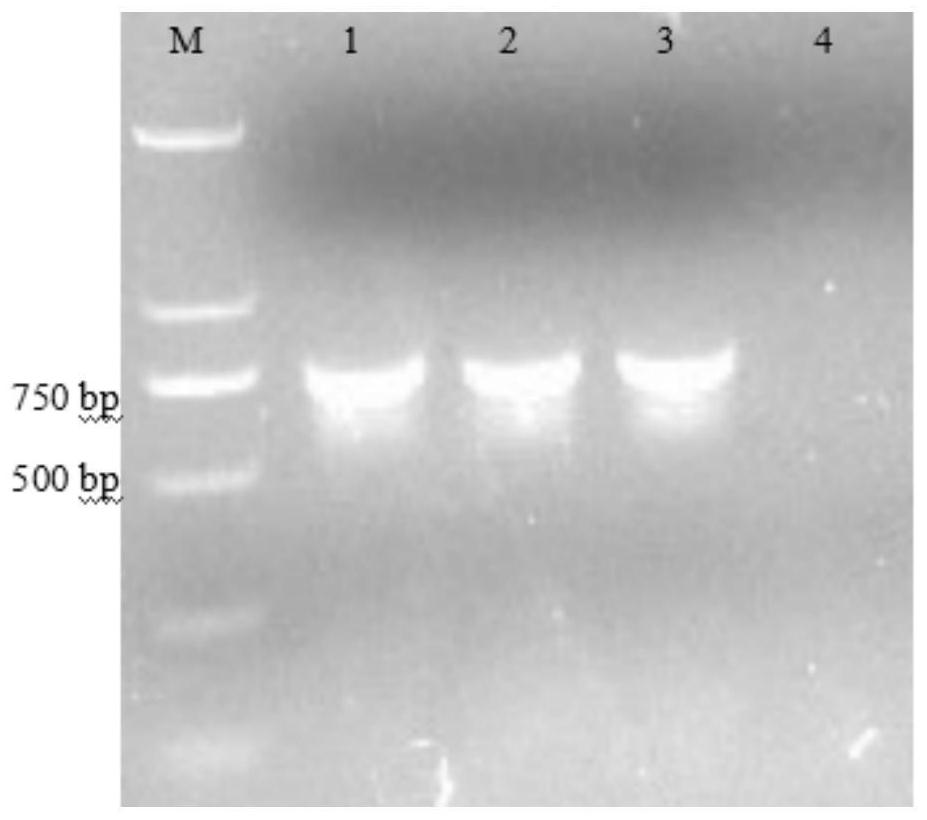
[0057] Using the standard strain of Escherichia coli ATCC 25922 as a positive control, Escherichia coli and its SCVs were detected for the amplification of specific gene phoA. The amplified product was 720 bp in size and a single bright band was positive for the amplification. This experiment uses 25μL PCR amplification system: dd H 2 O (20.35 μL), Easy Taq enzyme (0.15 μL), Easy Taq buffer (2.5 μL), dNTP (0.5 μL), 0.5 μL of upstream and downstream primers (see Table 1 for primer sequences), and 0.5 μL of DNA template. The conditions for PCR amplification of phoA gene were: pre-denaturation at 95°C for 5 min, denaturation at 95°C for 50 s, annealing at 55°C for 50 s, a total of 30 cycles, extension at 72°C for 1 min, and final extension at 72°C for 7 min.
[0058] Table 1 Primer sequences of phoA gene
[0059]
[0060]
[0061] figure 2 with Figure 7 Electropherograms for the detection of Escherichia...
Embodiment 3
[0063] Biochemical test, growth curve determination and auxotrophic test of Escherichia coli and its SCVs
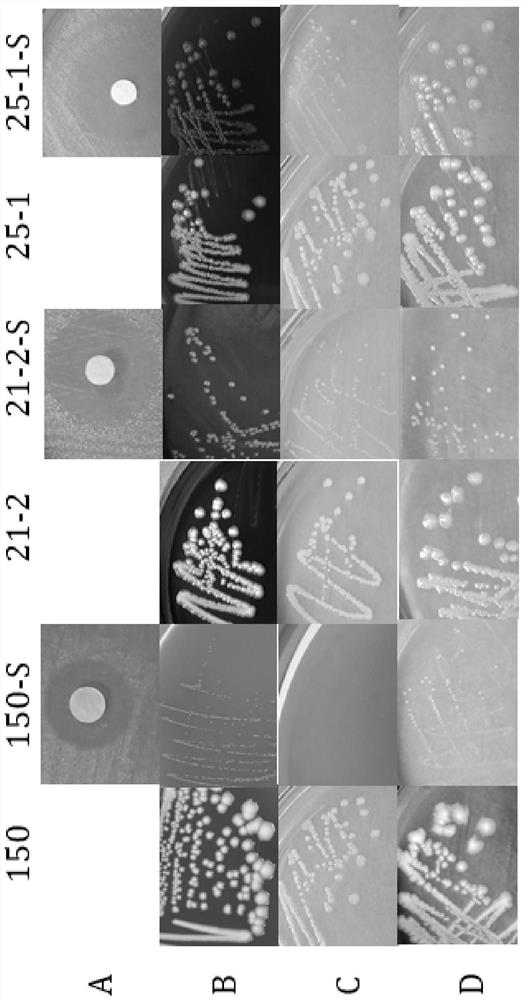
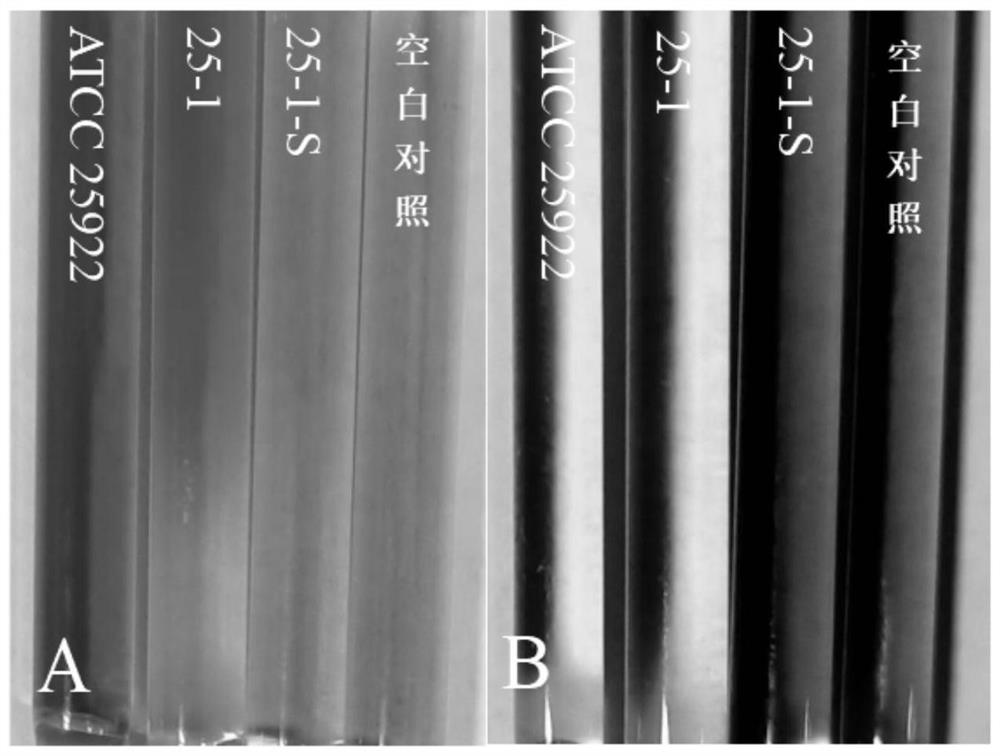
[0064] 1. With Escherichia coli standard strain ATCC 25922 and sterile normal saline as positive and blank controls respectively, use micro biochemical identification tubes to carry out sugar fermentation experiments on Escherichia coli and its SCVs (lactose, sorbitol, mannitol, glucose and trisaccharide iron ) and catalase test, nitrate reduction test and acetate utilization test.
[0065] Among them, in the acetate utilization test, the wild strains all utilized acetate, while the induced strains did not utilize acetate (results see image 3), the wild strain and its SCVs all fermented and utilized glucose and lactose, and the results are shown in Table 2. image 3 Among them, A is the acetate utilization test, if the strain to be tested can utilize acetate, it will appear blue. If the strain to be tested cannot utilize acetate, it will not change color or be light y...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com