Nucleic acid methylation cytosine conversion method
A technology for methylated cytosine and nucleic acid, applied in the field of nucleic acid methylated cytosine conversion, can solve the problems that hinder the commercial application of TAPS technology, difficulty in automation, DNA loss, etc., and achieve small DNA damage, convenient automation, and high conversion rate high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
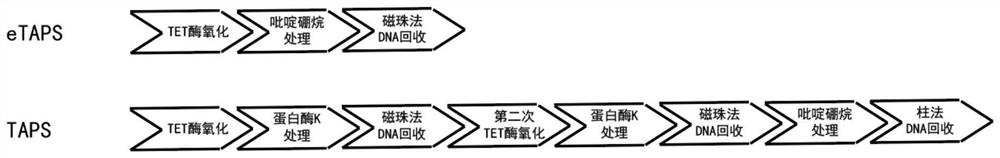
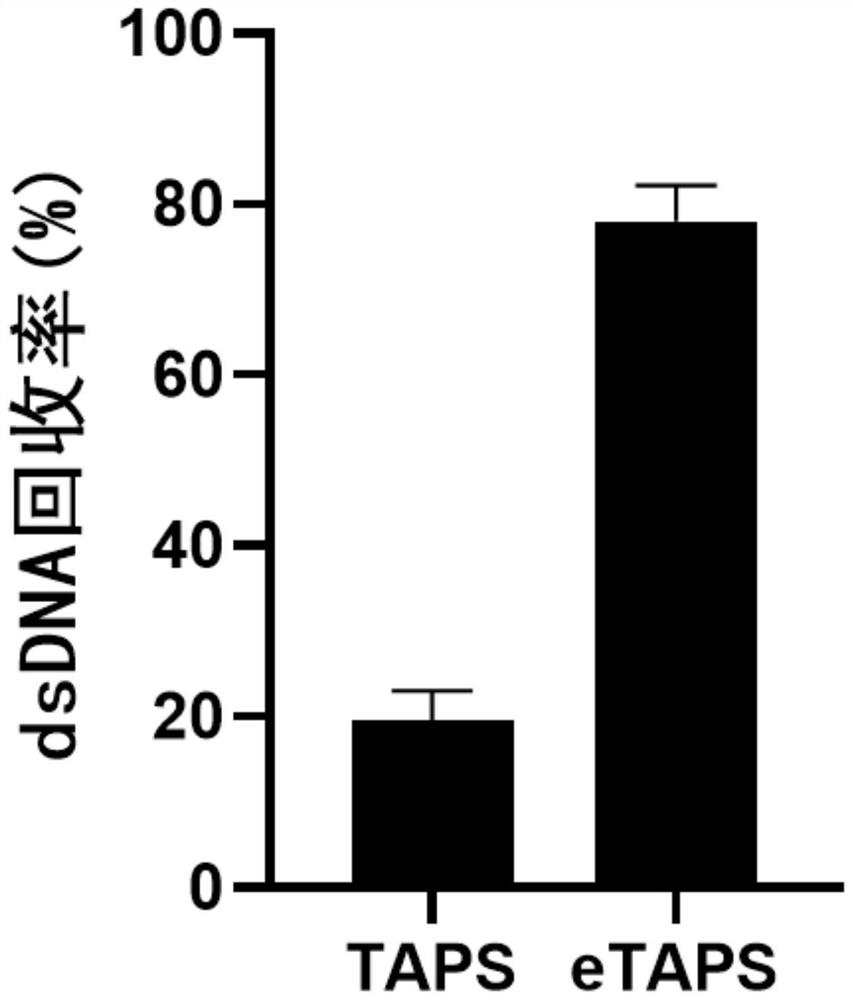
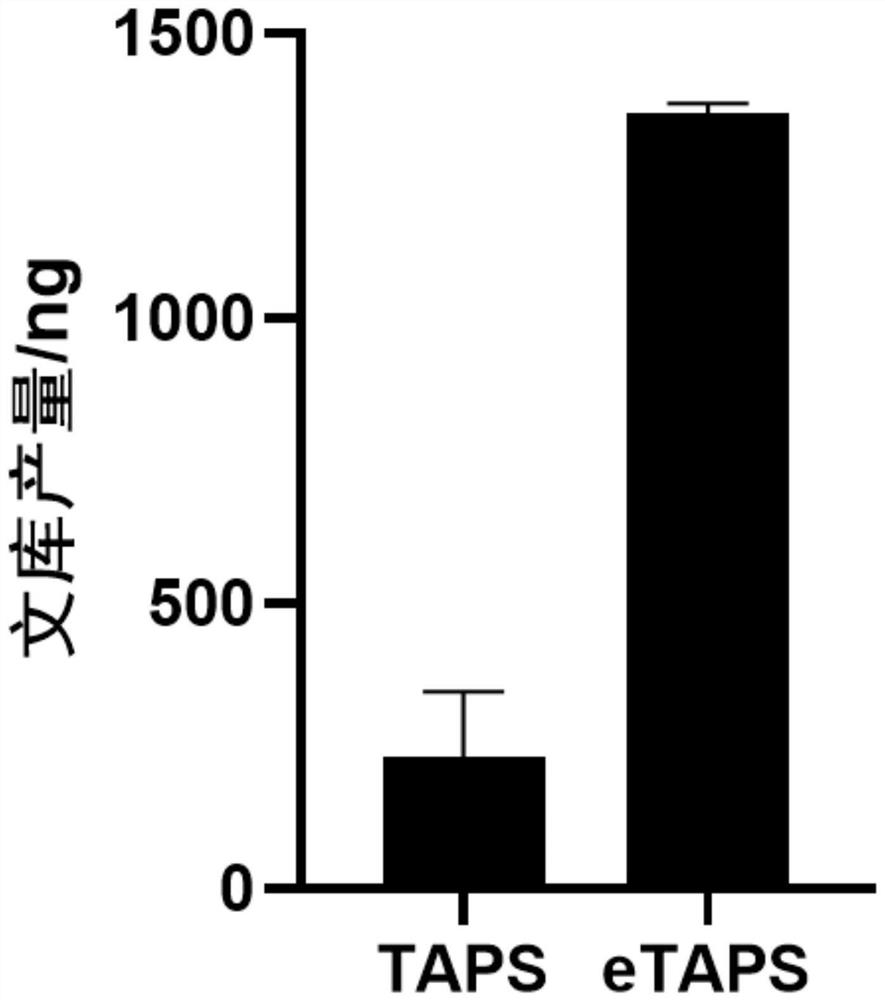
[0025] Embodiment 1: TAPS and eTAPS performance comparison
specific Embodiment approach
[0026] In this example, the conversion, DNA recovery and accuracy of TAPS and eTAPS were compared (see figure 1 ). The specific implementation is as follows:
[0027] 1) TAPS process:
[0028] In the first step, TET oxidation. Prepare the system according to the table below.
[0029] Table 1
[0030] components Dosage Methylated pUC19 DNA (Zymo Research, Cat#D5017) 10μL (10ng) 10× eTET1 Reaction Buffer 3μL 30× eTET1 auxiliary solution 1μL eTET1 (Yeasen, Cat#12219) 1μL H2O 15μL total 30μL
[0031] The 10× eTET1 reaction buffer included 500 mM tris, 10 mM α-ketoglutarate, 500 mM sodium chloride, and 2 mM dithiothreitol.
[0032] The 30× eTET1 supplement contains 3 mM ferrous ammonium sulfate and 60 mM vitamin C.
[0033] The second step is proteinase K treatment. Add 1 μL of 20 mg / mL proteinase K and react at 50°C for 0.5 h.
[0034] The third step is DNA recovery by magnetic bead method. Add 60 μL DNA Selected b...
Embodiment 2
[0057] Example 2: Application of eTAPS in the detection of RNA cytosine methylation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com