Multiplex RT-PCR method for simultaneously detecting four tobacco viruses
A technology of RT-PCR and tobacco virus, which is applied in the field of multiple RT-PCR, can solve the problems of cumbersome operation steps, cross-contamination, and long time consumption, and achieve the effects of improving detection efficiency, saving templates, and reducing detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
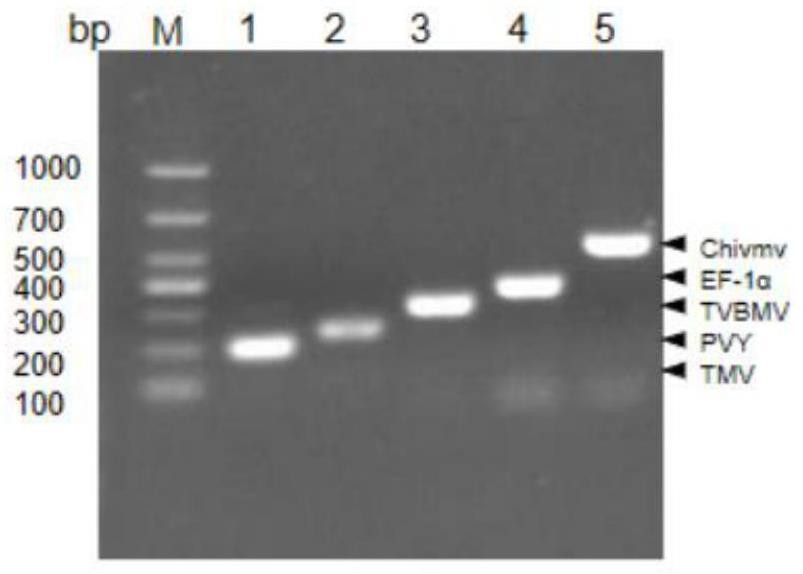
[0047]Primer design is carried out through the S2 step of the present invention. All primers of the present invention are designed after multiple alignments from the CP gene sequences of 4 viruses and an internal reference gene sequence. The sizes of the single polymerase chain reaction amplification products are respectively 201bp (TMV), 252bp (PVY), 362bp (EF-1α), 306bp (TVBMV) and 520bp (ChiVMV) expected fragments such as figure 1 shown. In the single polymerase chain reaction experiment, the cDNA mixture solution of 4 kinds of viruses was used as the template, but only the specific band added with a single virus primer pair was amplified in each reaction. This demonstrates the specificity of the designed primers and shows that these primer pairs can be successfully incorporated into multiplex reverse PCR.
Embodiment 2
[0049] The specific bands corresponding to each virus in the RT-PCR amplification products were gel-cut, recovered and purified with an ordinary agarose gel DNA recovery kit. The recovered RT-PCR product was connected with the vector pGD19-T according to the reaction system at 16°C for 2h 30min. Take the ligation product and transform it into E.coliDH5α competent cells, spread it on the LB medium plate containing Amp (100mg / mL), incubate at 37°C for 14-16h, carry out blue-white screening, and use colony PCR to screen the clone containing the insert , Inoculate the screened positive single colony into LB liquid medium containing Amp (100mg / mL), and culture overnight at 37°C. The plasmids of positive clones were extracted and sent to Sangon Biotech for sequencing with M13 forward and reverse primers (universal primers). These sequences are then verified by BLAST comparison of the NCBI nucleotide database; through the verification of the S4 step of the present invention, the seq...
Embodiment 3
[0051] Optimization of multiplex polymerase chain reactions;
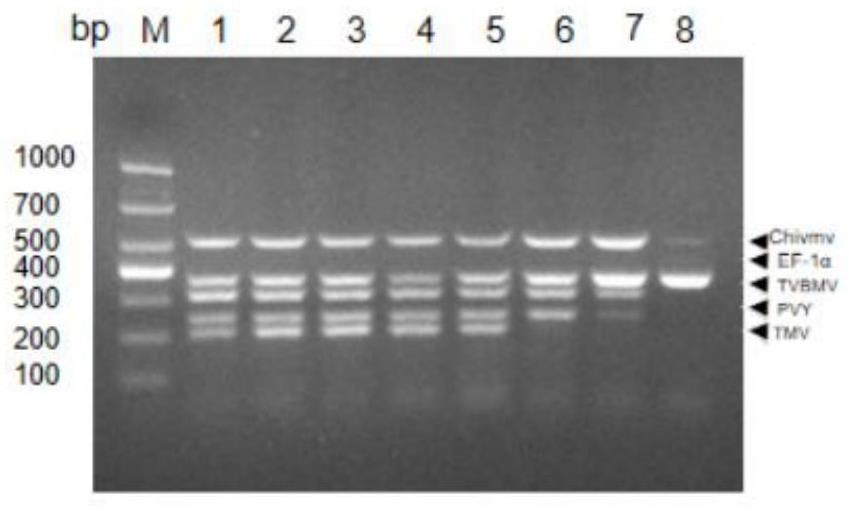
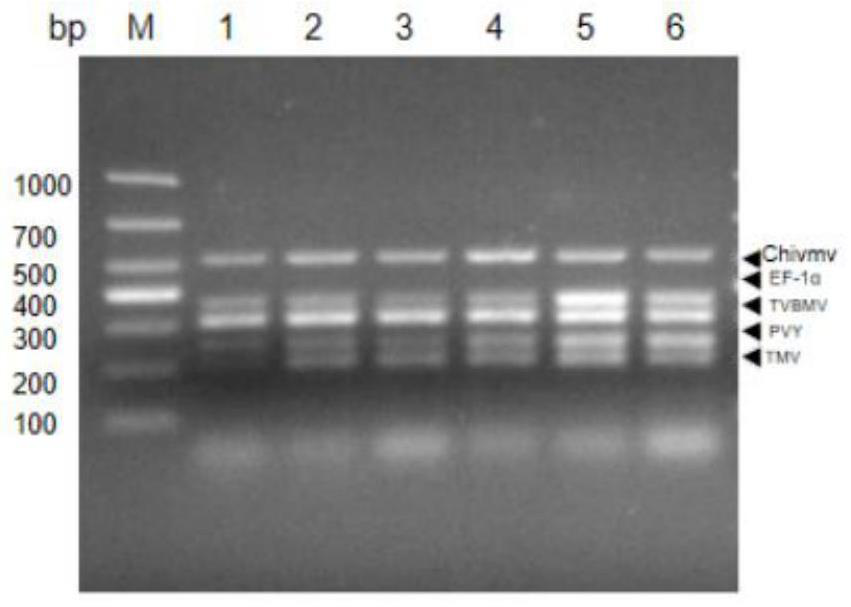
[0052] In order to determine the best combination of multiplex polymerase chain reaction, many factors were optimized for the multiplex system, including the ratio of primers, annealing temperature, annealing time, extension time and cycle time; 4 viral plasmids were used to mix plasmids (10pg ) was used as a template to optimize the multiple reverse transcription-polymerase chain reaction; first, 5 pairs of primers with different ratios were optimized, and finally the optimal primer volumes were determined to be 1.0 μL (TMV), 2.0 μL (ChiVMV), 0.5 μL (TVBMV), 0.5 μL (PVY), 1.0 μL (EF-1α); the optimized primer amount mixture was used for multiplex polymerase chain reaction; the annealing temperature was from 44°C to 65°C in increments of 3 for each gradient ℃; when the annealing temperature was 44-53℃, the brightness of TMV, ChiVMV, TVBMV, PVY and EF-1α target fragments increased with the increase of annealing tempe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com