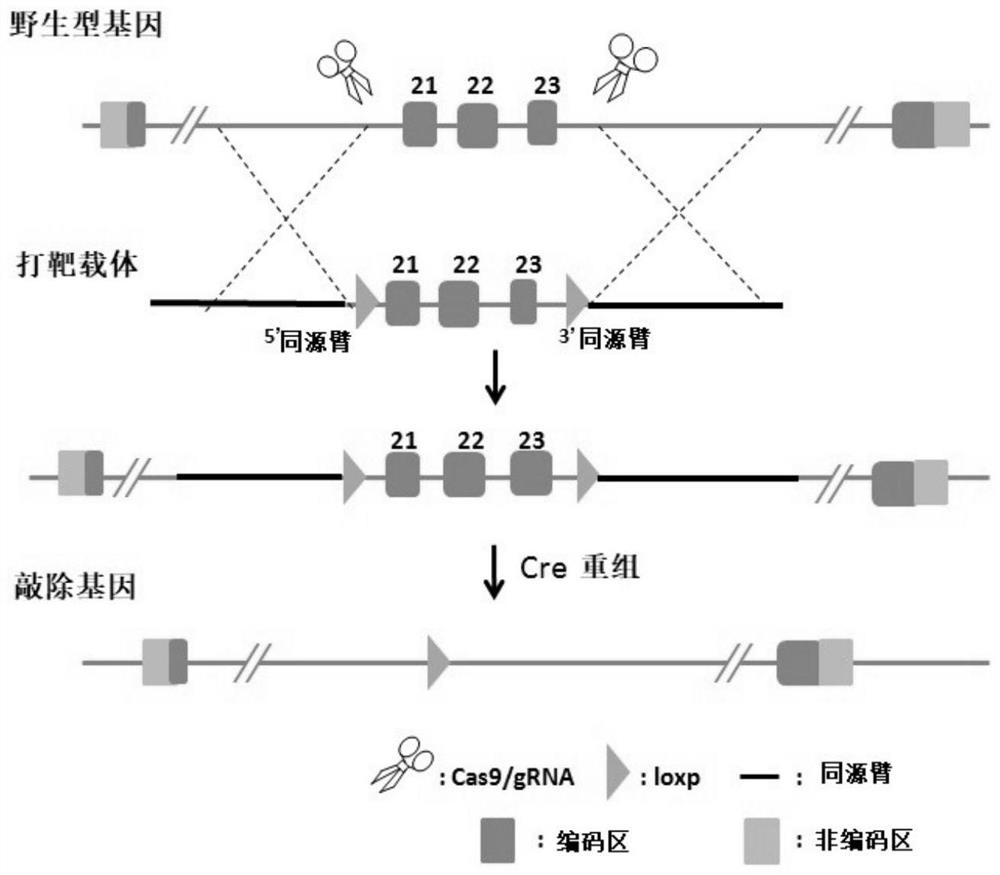
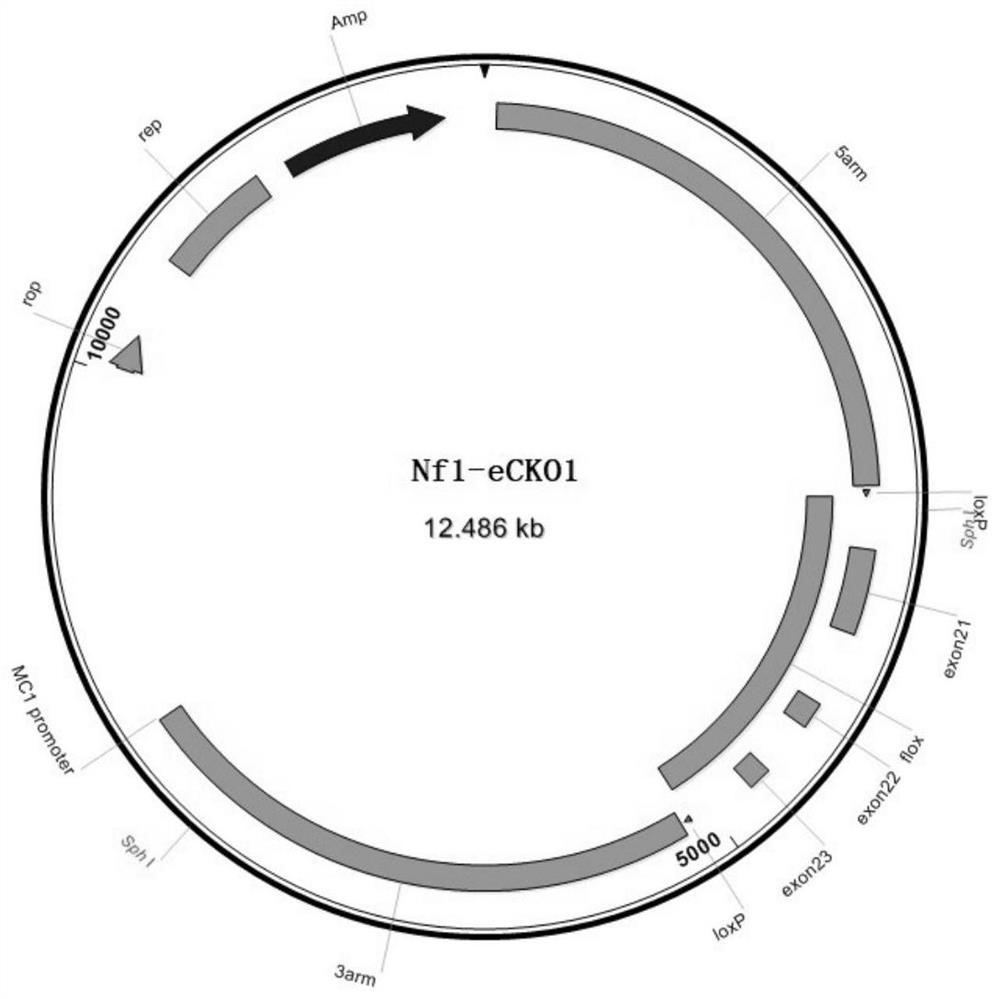
Construction method of Nf1 gene knockout animal model
A technology for constructing methods and animal models, applied in the field of gene editing, can solve the problems of difficult operation, low scalability, and high technical difficulty, and achieve the effect of good controllability, easy operation, and clinical characteristics.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Embodiment 1, obtaining gRNA
[0039] The method for obtaining the gRNA directed at the Nf1 gene of the present embodiment is as follows:
[0040] (1) Selection of cells used for gene targeting: Schwann cells of mice with C57BL / 6 genetic background;
[0041] (2) Design the gRNA target sequence for the Nf1 gene and use it to prepare gRNA primers: Primer01 is shown in SEQ ID NO.1 or SEQ ID NO.2, and Primer02 is shown in SEQ ID NO.3. Centrifuge Primer01 and Primer02 at 13,000 rpm for 1 min. After completion, add 75 μL ddH2O to the Primer01 tube, add 256 μL ddH2O to the Primer02 tube, and directly stand at room temperature for later use;
[0042] (3) Prepare a mixed system in a PCR tube according to the following system:
[0043]
[0044] ( Max DNA Polymerase (Cat. No.: R045A)
[0045] (4) 150 μL system was divided into six tubes, each tube was 25 μL. The PCR reaction was carried out in a PCR machine, and the program was as follows: 98°C, 2min denaturation, 98°C fo...
Embodiment 2
[0048] Embodiment two, obtain Cas9 mRNA
[0049] In this example, Cas9 mRNA was obtained by in vitro transcription. The specific method is as follows:
[0050] (1) 10 μg of plasmid pX-T7 (Cas9), 5 μL of XbaI enzyme, and 100 μL of the total system; linearized with XbaI, and reacted at 37°C for 2 hours;
[0051] (2) Add 10 μL ammonium acetate, mix well, add 200 μL absolute ethanol, mix well, centrifuge at 13000 rpm for 5 min, discard the supernatant, centrifuge at 13000 rpm for 1 min, blot the residual liquid, add 20 μL nuclease-free water;
[0052] (3) In vitro transcription using mMESSAGE mMACHINE T7 ULTRA Kit (Invitrogen, AM1345);
[0053] (4) Use MEGAclear TM Transcription Clean-Up Kit (Invitrogen, AM1908) was purified and stored at -80°C.
Embodiment 3
[0054] Example 3, construction of homologous recombination vector
[0055] The construction method of the homologous recombination vector of this embodiment is as follows:
[0056] (1) Use the sequences shown in SEQ ID NO.8 and SEQ ID NO.9 as primers, use the wild-type mouse genome as a template, and obtain the target fragment of the 5' homology arm by PCR amplification; use such as SEQ ID NO. The sequences shown in ID NO.10 and SEQ ID NO.11 are used as primers, and the wild-type mouse genome is used as a template to perform PCR amplification to obtain the target fragment of the flox region; using such as SEQ ID NO.12 and SEQ ID NO. The sequence shown in 13 was used as a primer, and the wild-type mouse genome was used as a template to obtain the target fragment of the 3' homology arm by PCR amplification. The PCR primers are shown in Table 1:
[0057] Table 1
[0058]
[0059] (2) Digest the plasmid pBR-322 with BamHI and HindIII to obtain the linearized pBR-322 backbone ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com