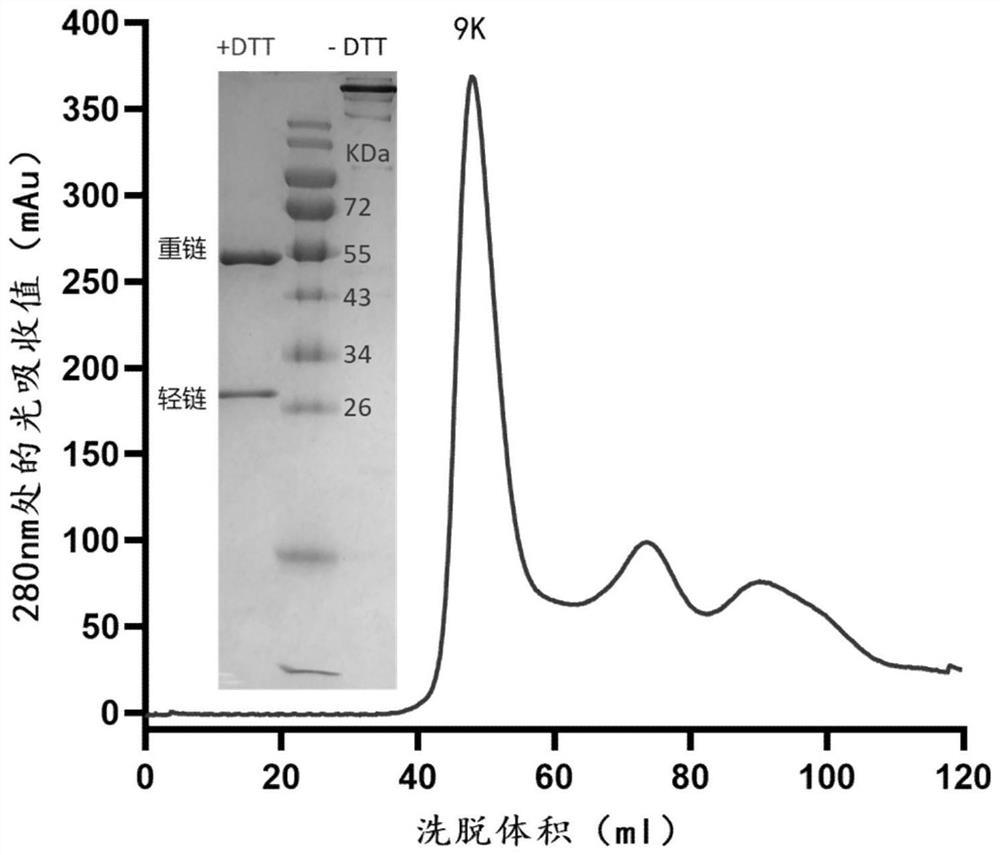
Novel coronavirus, monoclonal antibody of mutant of novel coronavirus and application of monoclonal antibody
A monoclonal antibody and antigen technology, applied in the fields of immunology and molecular virology, can solve problems affecting the effect of neutralizing antibodies, and achieve the effect of clinical application value in prevention and treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
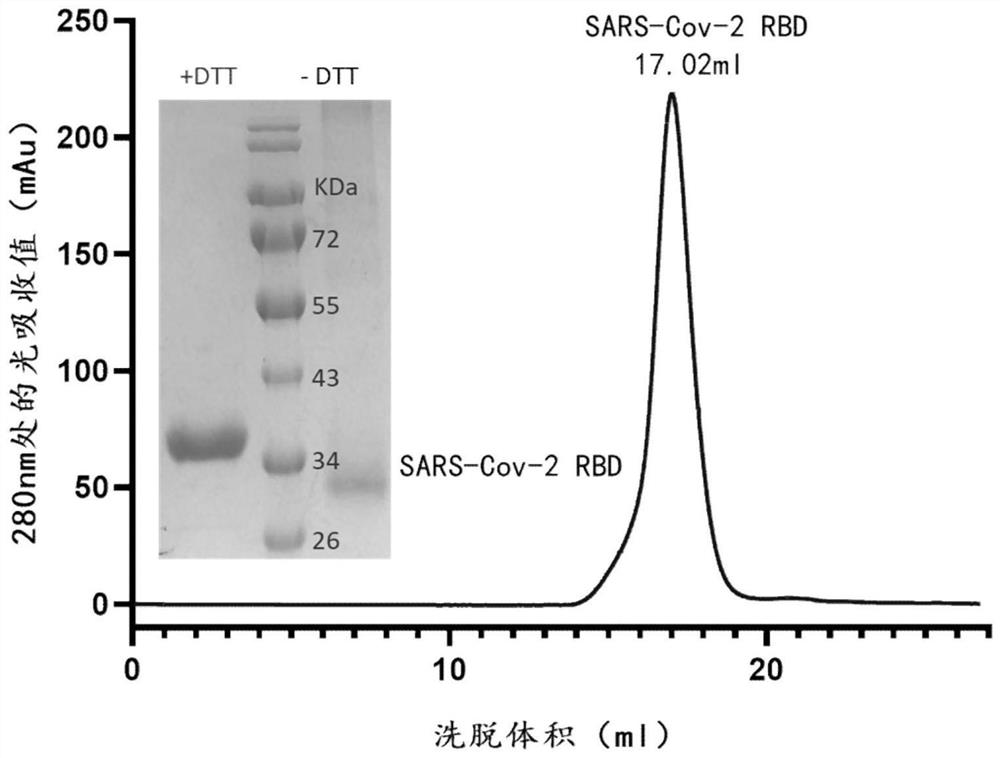
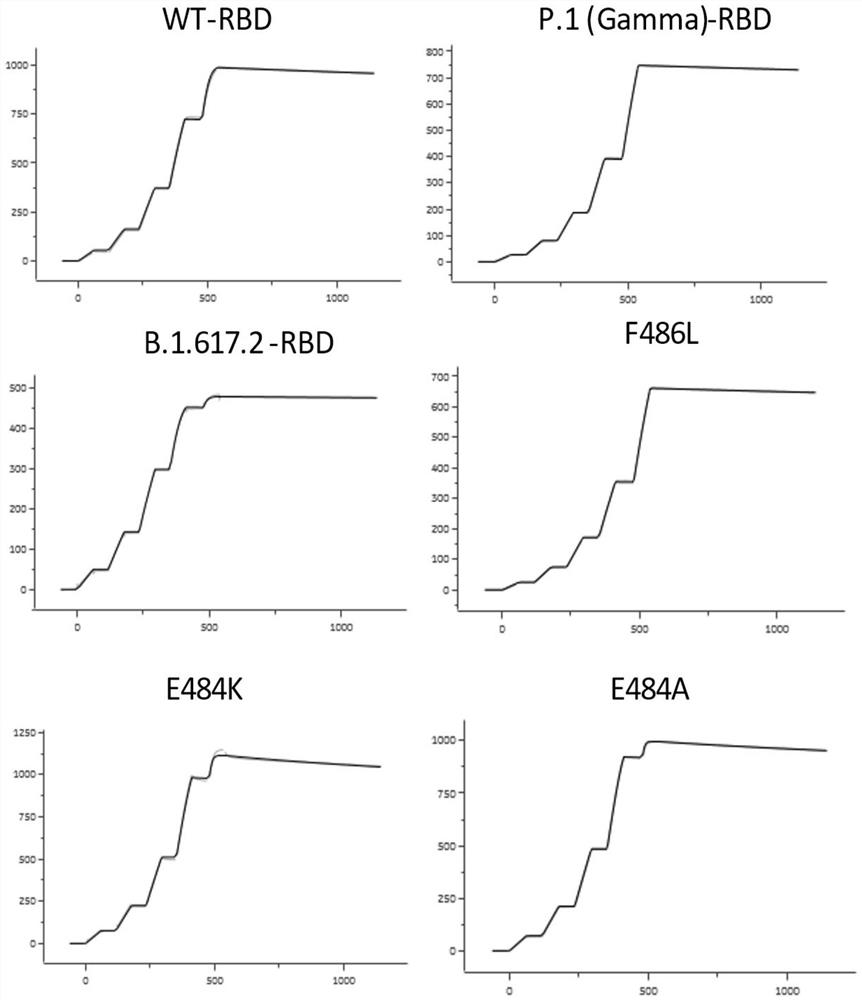
[0100] Expression and purification of embodiment 1SARS-CoV-2 virus S protein RBD
[0101] The optimized wild-type nCoV-RBD (residues 319-541, GenBank: YP_009724390.1) coding sequence with 6 His tags at the C-terminus was cloned into the mammalian expression vector pCAGGS. The various mutant RBDs shown in Table 1 (K417N, K417T (present in P.1 (Gamma) RBD), L452R (present in B.1.617.1 (Kappa) RBD and B.1.617.2 (Delta ) RBD), Y453F, N460S, T478K (present in B.1.617.2 (Delta) RBD), E484K, E484A, F486L, N501Y (present in B.1.1.7 (Alpha) RBD, B.1.351 (Beta) ) RBD, P.1 (Gamma) RBD), N501T) coding genes were subcloned into pCAGGS. Then the plasmid (2 μg) and PEI were transiently co-transfected at a mass ratio of 1:3 per milliliter of HEK293F cells. At 310K, 5% CO 2 The cells were cultured with SMM 293-TII medium (Sino Biological) under certain conditions, and then supplemented with SMS M293-SUPI (Sino Biological) at a ratio of 35 mL / L 24 hours after transfection. On the fifth day,...
Embodiment 2
[0102] Example 2 Isolation of memory B cells that specifically recognize RBD protein
[0103] With the informed consent of those infected with the SARS-CoV-2 virus who were cured and discharged, 10 mL of blood was collected and PBMCs were isolated. Separated PBMCs in 10 7 / mL density and final concentration of 400nM RBD protein (prepared by Example 1) were incubated on ice for half an hour; then washed twice with PBS, and then incubated with the following antibodies (both purchased from BD): anti-human CD3 / PE-Cy5, anti-human CD16 / PE-Cy5, anti-human CD235a / PE-Cy5, anti-human CD19 / APC-Cy7, anti-human CD27 / Pacific Blue, anti-human CD38 / APC, anti- human IgG / FITC, and anti-His / PE. After incubation on ice for half an hour, wash PBMCs twice with PBS. Subsequently, sort PBMCs with FACSAria III and collect PE - Cy5 - APCs - APCs - Cy7 + Pacific Blue + FITC + PE + The cells (that is, B cells) were directly collected into a 96-well plate, 1 cell / well.
Embodiment 3
[0104] Example 3 Isolation and identification of 9K antibody and construction of recombinant expression vector
[0105] The B cells obtained in Example 2 were reverse-transcribed using Superscript III reverse transcriptase (Invitrogen) (at 55° C. for 60 minutes), wherein the reverse transcription primers used are shown in Table 2.
[0106] Table 2 Sequence information of reverse transcription primers used
[0107]
[0108]
[0109] Using the reverse transcription product as a template, the first round of PCR (PCRa) was carried out with HotStar Tap Plus enzyme (QIAgen) to amplify the sequence of the variable region of the antibody; wherein, the primers used are shown in Table 3; the reaction conditions used As follows: 95°C, 5min; 35 cycles of (95°C for 30s, 55°C (heavy chain / κ chain) for 30s, 72°C for 90s); 72°C, 7min. Subsequently, the second round of PCR (PCRb) was carried out using the amplified product as a template; wherein, the primers used were as shown in Table ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com