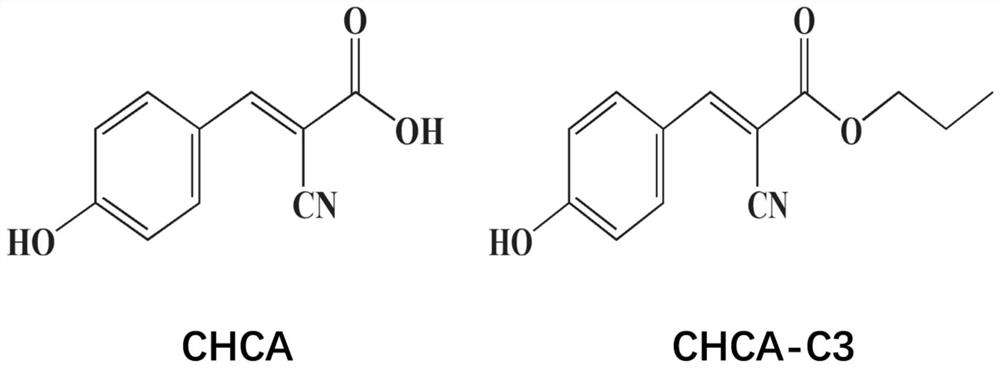
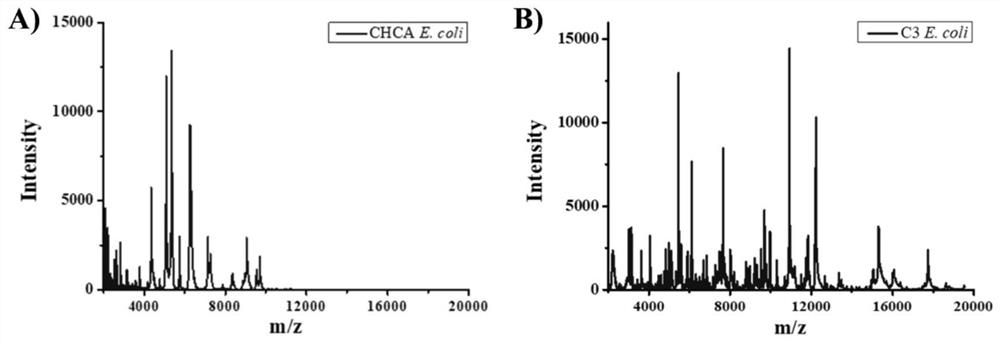
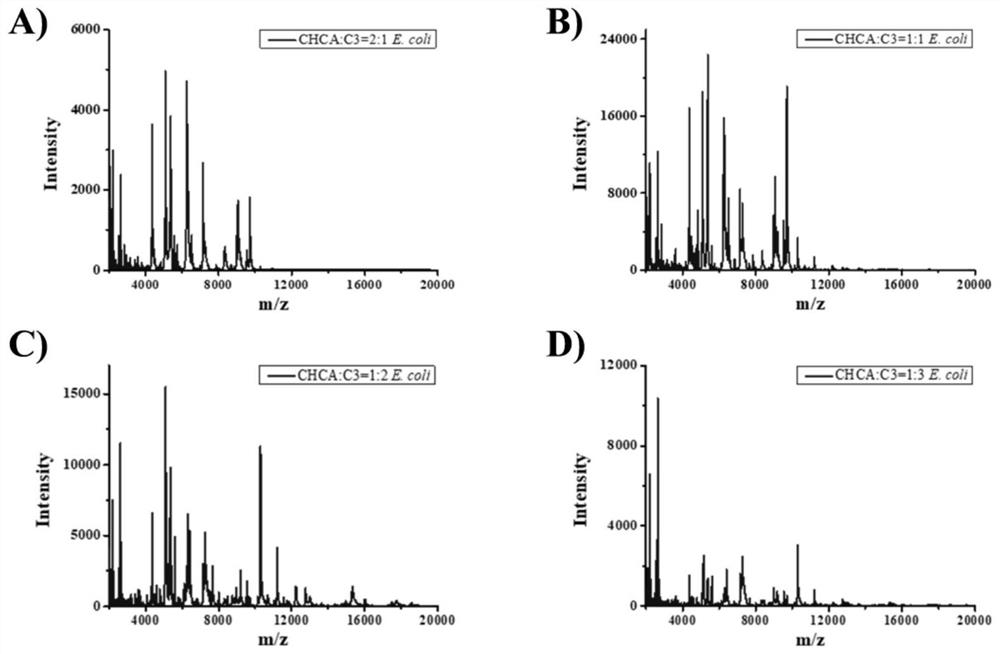
Application of novel mixed matrix in MALDI-MS (matrix-assisted laser desorption ionization-mass spectrometry) bacterial identification
A mixed matrix, bacterial identification technology, applied in the direction of resisting vector-borne diseases, instruments, measuring devices, etc., can solve the problems of inability to distinguish similar bacteria, incomplete fingerprints, etc., and achieve accurate and highly sensitive identification of clinical drug delivery. , the effect of expanding the number of mass spectral peaks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] The step of detecting bacterial mixed matrix in MALDI mass spectrometry as follows:
[0078] Step a: cultured on solid medium were Pseudomonas aeruginosa and Enterobacter 24h;
[0079] Step 2: CHCA and CHCA-C3 were formulated in 50% acetonitrile containing 2.5% trifluoroacetic acid solution in a concentration of 10-15mg / mlCHCA CHCA-C3 substrate solution and stored at 4 ℃ refrigerator to standby;
[0080] Step Three: CHCA matrix solution were taken and CHCA-C3 substrate solution at a volume ratio of 1: 2 were uniformly mixed;
[0081] Step Four: a culture from step Enterobacter and Pseudomonas aeruginosa were obtained uniformly coated on a supporting MALDI-MS target plate;
[0082] Step Five: Enterobacter and Pseudomonas aeruginosa on Pseudomonas Step Four target plates after air-drying, the mixed matrix solution obtained in step three points each were taken 0.5-1μL Pseudomonas aeruginosa and Enterobacter on the dry ;
[0083] Step Six: A solution of step 5 was added dropw...
Embodiment 2
[0087] Mixed matrix determination step Shigella and E. coli in MALDI mass spectrometry as follows:
[0088] Step a: On solid medium were cultivated Escherichia coli and Shigella 24h;
[0089] Step 2: CHCA and CHCA-C3 were formulated in 50% acetonitrile containing 2.5% trifluoroacetic acid solution in a concentration of 10-15mg / mlCHCA CHCA-C3 substrate solution and stored at 4 ℃ refrigerator to standby;
[0090] Step Three: CHCA matrix solution were taken and CHCA-C3 substrate solution at a volume ratio of 1: 2 were uniformly mixed;
[0091] Step Four: obtained from step a culture of Escherichia coli and Shigella are uniformly coated on a supporting MALDI-MS target plate;
[0092] Step Five: after Step Four target plates on Escherichia coli and Shigella air drying, the mixture obtained in step three matrix solution were taken on 0.5-1μL point after dry Escherichia coli and Shigella;
[0093] Step Six: Escherichia coli step 5 was added dropwise a mixed solution of the matrix and S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com