Methods for identifying activating antigen receptor (ACAR)/inhibitory chimeric antigen receptor (ICAR) pairs for use in cancer therapies
A chimeric antigen receptor, inhibitory technology for the identification of activating antigen receptor (aCAR)/inhibitory chimeric antigen receptor (iCAR) pairs for cancer therapy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0570] vii. Preparation of target cells
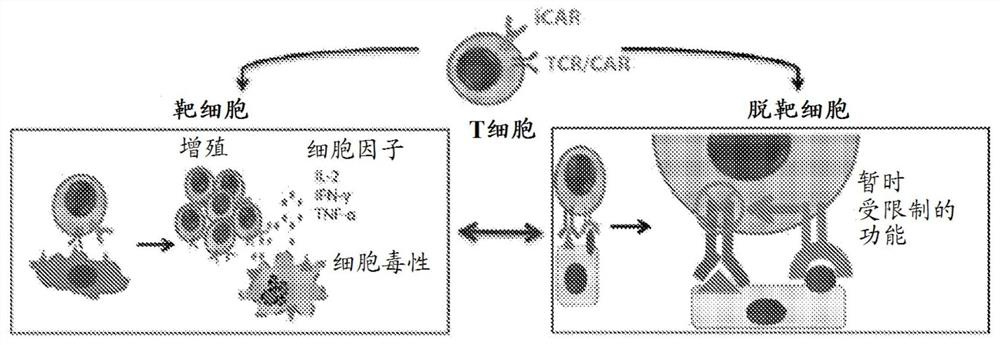
[0571] In some embodiments, target cells are prepared and tested in an in vitro system. In some embodiments, an in vitro recombination system will be established for testing the function of the iCAR and / or pCAR constructs in inhibiting aCAR activity against off-target cells. In some embodiments, target cells expressing aCAR epitopes, iCAR epitopes, or both will be generated. In some embodiments, target cells expressing aCAR epitopes, pCAR epitopes, or both will be generated. In some embodiments, recombinant cells expressing aCAR epitopes will represent on-tumor ('on-tumor') cells on target, while cells expressing aCAR and iCAR epitopes will represent on-tumor ('on-tumor') on-target cells. 'off-tumor') healthy cells.
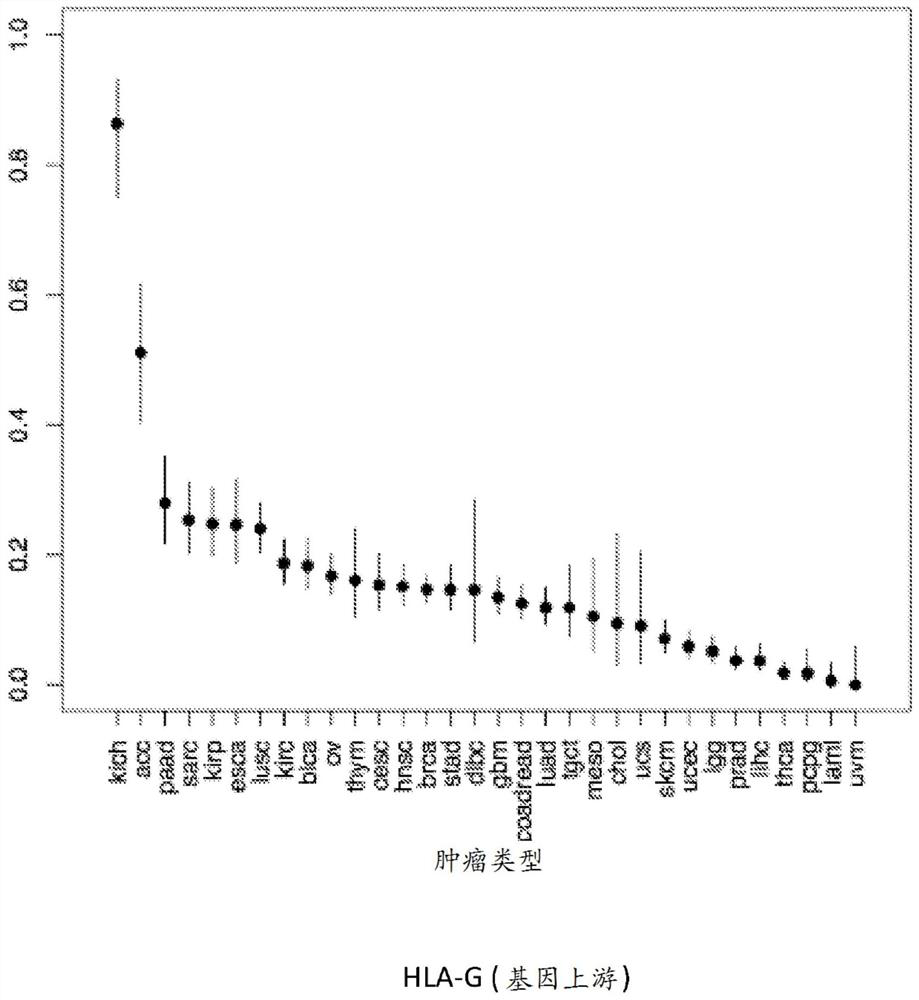
[0572]In some embodiments, the iCAR / aCAR panel will be HLA (including, for example, HLA-A2, HLA-A3, HLA-A, HLA-B, HLA-C, HLA-G, HLA-E, HLA-F, HLA- DPA1, HLA-DQA1, HLA-DQB1, HLA-DQB2, HLA-DRB1 or HLA-DRB5) and CD19. I...
Embodiment 1
[0865] Example 1. LOH rate assessment of HLA genes across cancers
[0866] Introduction:
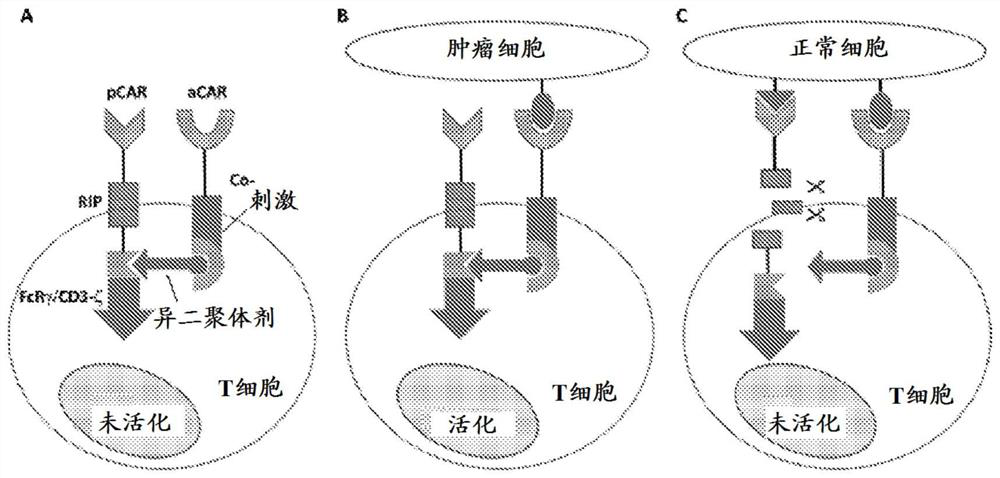
[0867] A therapeutic strategy is proposed to address the vulnerability caused by genomic loss in cancer cells. The proposed strategy uses a combination of activating CAR T cells (aCAR) and inhibitory CAR T cells (iCAR) to more safely target heterozygous cells that have lost encoding membrane proteins for both maternal and paternal alleles. Tumors with combined genomic segments (i.e., polymorphic protein-coding changes).
[0868] If the target of iCAR is only expressed by non-tumor tissues, iCAR can reduce the off-tumor toxicity of CAR-T therapy without reducing the anti-tumor efficacy. This situation, in which the iCAR target is only expressed by non-tumor cells, occurs when the iCAR antigen is encoded by a part of the genome that has been deleted in tumor cells. One gene family that is highly polymorphic and known to be expressed on all cells is the HLA.
[0869] HLA proteins are al...
Embodiment 2
[0903] Example 2. Genome-wide identification of germline alleles encoding expressed cell surface proteins undergoing loss of heterozygosity
[0904] Introduction:
[0905] If the target of iCAR is only expressed by non-tumor tissues, suppressive CAR-T cells can reduce the off-tumor toxicity of CAR-T therapy without reducing the anti-tumor efficacy. The situation where the iCAR target is only expressed by non-tumor cells is where the iCAR antigen is encoded by a part of the genome that has been deleted in the tumor cells. The goal of this part of the workflow is to identify such alleles.
[0906] Allele Identification:
[0907] We used the Exome Integration Consortium (ExAC) database as input for the analysis (exac.broadinstitute.org). The ExAC database is a compilation of exomes from various population-level sequencing studies, totaling 60,706 exomes 1. ExAC contains information about each variant, including the number of reference allele counts (allele frequency) compare...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com