Tumor neoantigen prediction method based on HPV integration
A prediction method, antigen technology, applied in the field of bioinformatics, can solve the problems of deletion, chromosome amplification, host cell genome instability, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
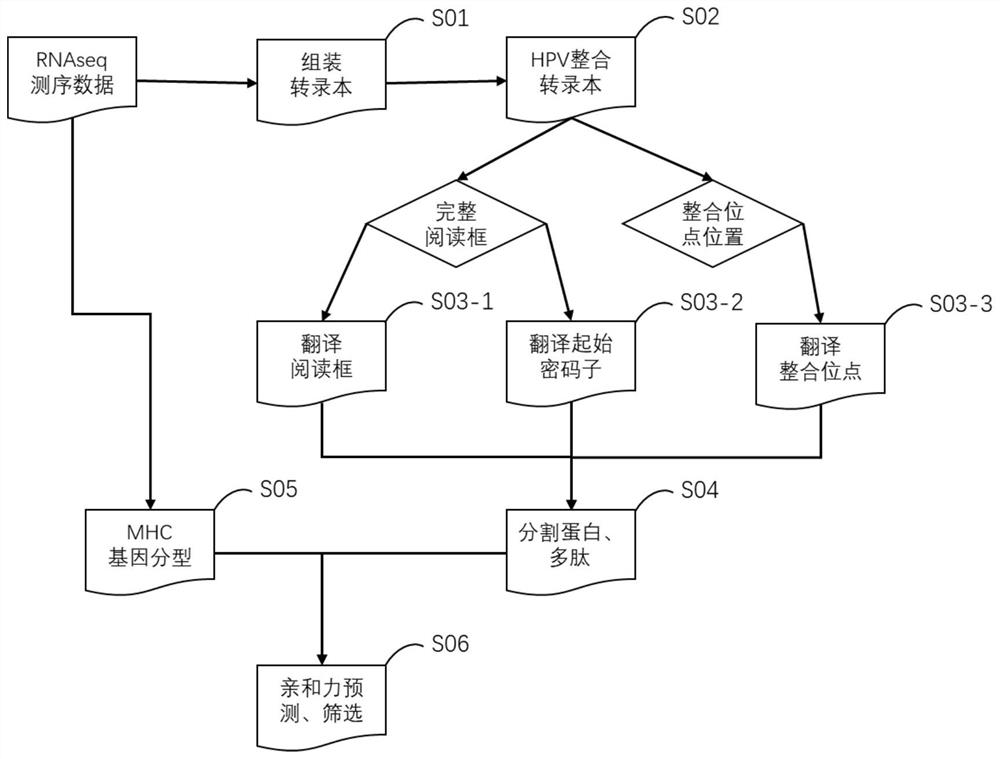
[0039] Such as figure 1 Shown, a kind of neoantigen prediction method based on HPV integration, the method comprises the following steps carried out by processor:
[0040] S01, Assembly of tumor sample transcripts
[0041] Specifically, firstly, the ribosomal chain specific library construction method and the small fragment enrichment screening library construction method were used to construct the library and sequence, and obtain the deep sequencing data of the sample mRNA. The sample data includes multiple overlapping or partially overlapping short-read sequences. The degree of overlap is related to the depth of sequencing. The RNA-Seq sequencing data of a sample of not less than 15G should be obtained.
[0042] Secondly, filter the sequencing data, remove short-read sequences with an average base quality of less than 20 or contain sequencing primer adapters in the sequencing data, and convert the data format into a form that can be accepted by the subsequent assembly softw...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com