Fusion protein and application thereof
A fusion protein and protein technology, applied in fusion protein and its application field, can solve problems affecting protein-DNA interaction, gene silencing, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0183] Example 1: Construction of demethylation gene editing tool vector
[0184] 1.1dCas9-TET1cd demethylation tool
[0185] (1) Use the existing dCas9 and TET1 sequences in the laboratory, and use high-fidelity enzyme Q5 to amplify the sequence fragments of dCas9 and TET1cd. Gel recovered fragments for future use.
[0186] (2) Using the p1300-UBQ-CAS9 vector cut with Nco I and BamH I, the p1300-UBQ fragment was recovered by cutting the gel and used for future use.
[0187] (3) Using recombinase to recombine the dCas9 fragment into the p1300-UBQ fragment to obtain p1300-UBQ-dCas9, which is ready for use. Sanger sequencing was used to prove that the fragment recombination was successful.
[0188] (4) Then use BamHI to cut the p1300-UBQ-dCas9 vector obtained above.
[0189] (5) Using recombinase to recombine the TET1cd fragment into the p1300-UBQ-dCas9 fragment to obtain the p1300-UBQ-dCas9-TET1cd vector, which is the final vector for targeted editing of DNA methylation. S...
Embodiment 2
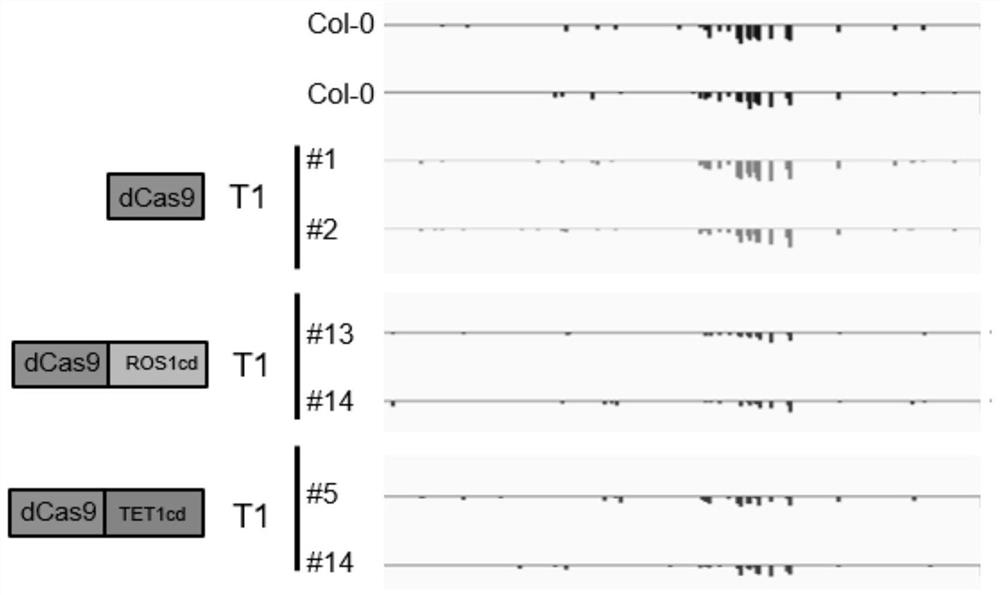
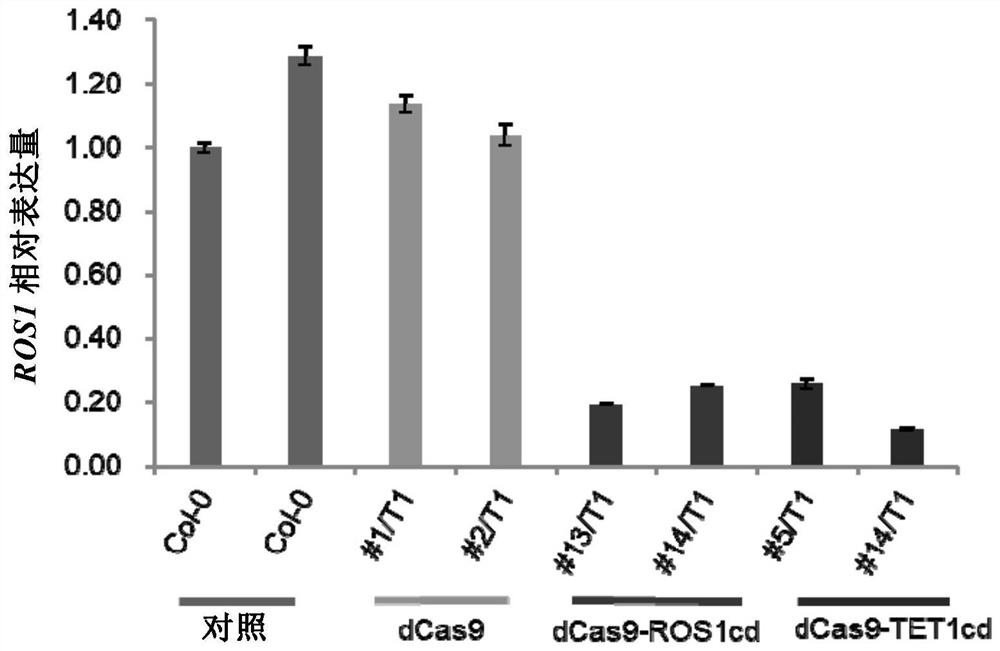
[0193] Example 2: Regulation of ROS1 expression by MEMS demethylation in the ROS1 promoter region
[0194] 2.1 Target design and construction
[0195] (1) Five sgRNAs targeting MEMS regions were designed according to the rules of sgRNA design, and the sequences of corresponding sgRNAs are shown in Table 1. In addition to the 20 bp targeting the MEMS region, the sgRNA also has a sticky end for ligation.
[0196] Table 1 sgRNA sequences targeting MEMS regions
[0197]
[0198]
[0199] (2) Change the F and R sequences of sgRNA into double-stranded DNA fragments with cohesive ends through annealling program. The process is as follows: dilute the forward and reverse primers to 100 μM, mix 1 μL each with 1 μL T4 DNA ligase buffer, 0.5 μL T4 polynucleotide kinase, and 6.5 μL ddH2O, keep the mixture at 37 °C for 30 min, 95 °C for 5 min, and then The speed of 0.2°C / s is reduced to 25°C, and finally diluted 250 times with water.
[0200] (3) U6, U3b, and 7SL vectors were dige...
Embodiment 3
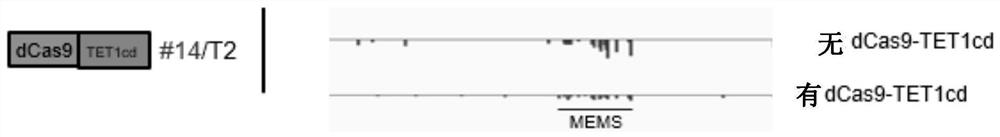
[0251] Example 3: Demethylation experiment in RdDM mutant (nrpd1)
[0252] 3.1 Target design
[0253] The target design is consistent with the previous ones. sgRNAs 1, 2, and 3 are connected to the upstream of the fusion protein, while sgRNAs 4, 5, and 6 are connected to the downstream of the fusion protein. The sequence of the sgRNA is shown in Table 3. The composition of the sgRNA sequence is the same as that of the previous one. sgRNAs are consistent.
[0254] Table 3 sgRNA sequences targeting multiple regions
[0255]
[0256]
[0257] 3.2 Genetic Transformation
[0258] See step 2 in the experimental example for the genetic transformation process.
[0259] 3.3 Screening of positive seedlings
[0260] Refer to step 3 positive seedling screening in the experimental example.
[0261] 3.4 Detection of DNA methylation level and editing efficiency
[0262] (1) When the positive seedlings grow to an appropriate size, the leaves of the positive seedlings are taken to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com