Artificial neuron calculation model construction method based on DNA cage structure
A neuron model and neuron computing technology, applied in the field of biological computing, can solve the problems of large number of DNA chains, cumbersome and complicated reaction process, leakage, etc., and achieve the effect of structural stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
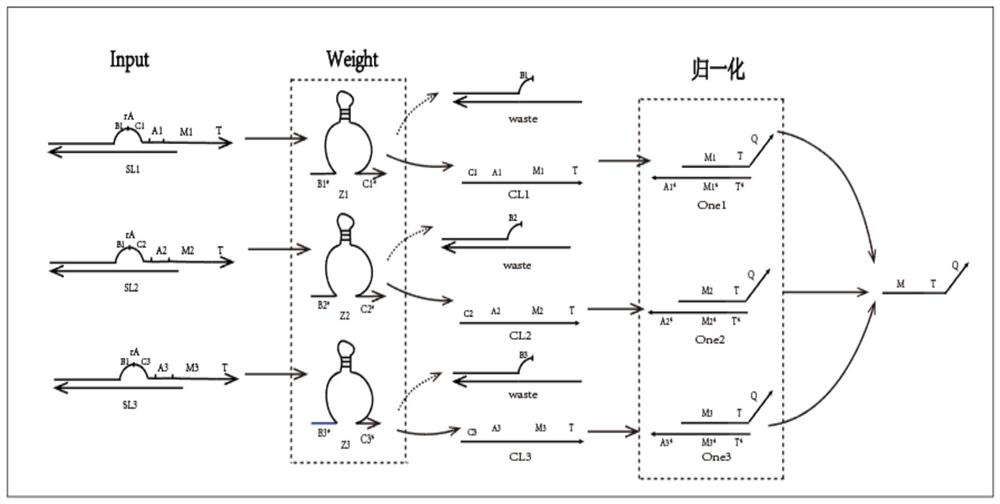
[0049] Example 1: Normalization module
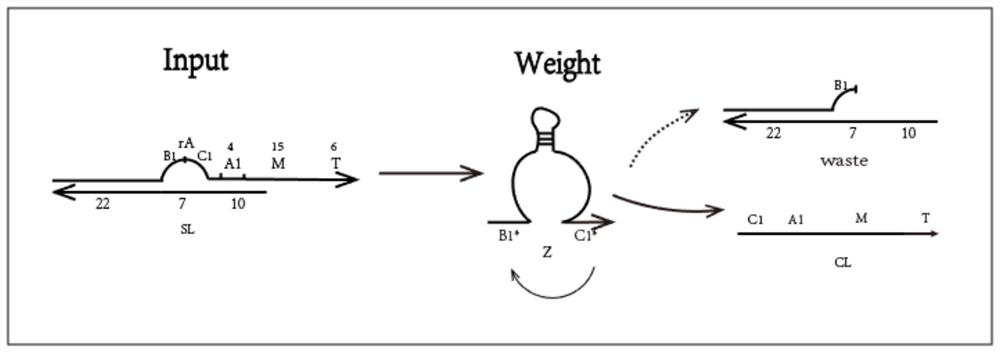
[0050] (1) Mix the rA-modified L1 chain (64 bases) with the small short chain S1 (39 bases) at a ratio of 1:1 in 1×TAE / Mg2+ buffer, and anneal from 90°C to At room temperature, a double-strand structure with rA-modified "bubbling" is formed, and the same is true for the other two double-strands SL2 and SL3; the normalization gates One1, One2, and One3 respectively divide the MTQ chain and the CL* chain at a ratio of 1:1. The ratio is mixed in 1×TAE / Mg2+ buffer and annealed from 90°C to room temperature to form a One gate complex;
[0051] (2) Put the input strands SL1, SL2, SL3, E6DNAzyme Z1, Z2, Z3 and each One gate into the same test tube with a concentration of 0.6uM, put them in an environment of 25°C, and react for about 12 hours;
[0052] (3) Figure 6 yes figure 2 The gel electrophoresis image of the corresponding product in (partial reaction image of the normalization module), lane 1 is the One1 gate, lane 3 is the CL1 chain...
Embodiment 2
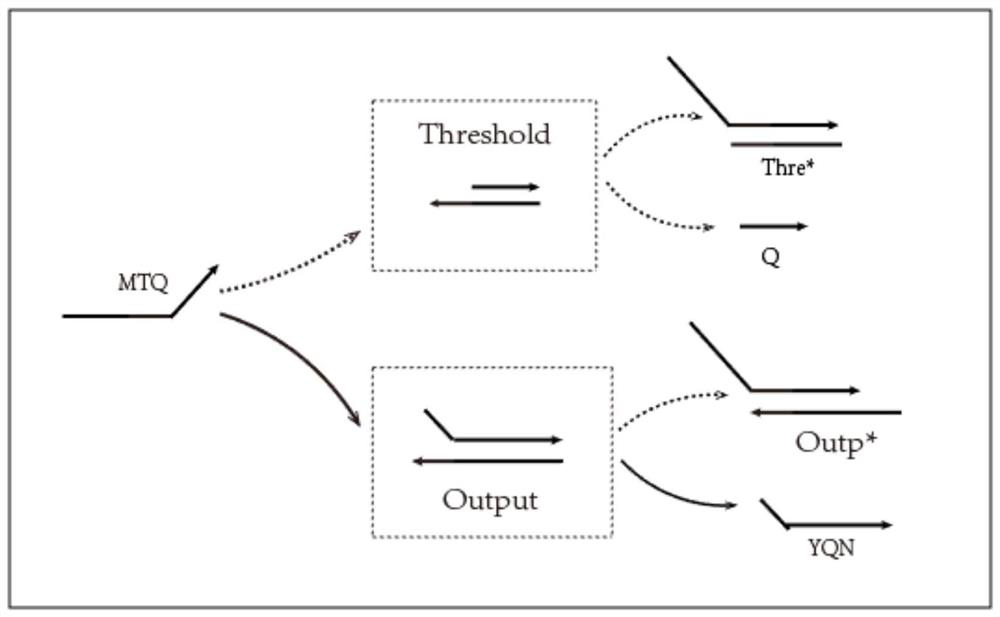
[0053] Embodiment 2: threshold function module
[0054] (1) Mix Q chain and Q*T*M_* chain in 1×TAE / Mg2+ buffer at a ratio of 1:1, and anneal from 90°C to room temperature to form Thre threshold gate complex; similarly, YQN chain and N*Q*T* chain are mixed in 1×TAE / Mg2+ buffer at a ratio of 1:1, and annealed from 90°C to room temperature to form an Outp gate complex;
[0055] (2) Place the MTQ chain and threshold gate Thre gate, MTQ chain and Outp gate in the ratio of 1:1 in test tube 4 and test tube 5 respectively, the concentration is 0.6uM; Put it in test tube 9 and test tube 10 according to the ratio of 1:1:1 and 2:1:1, the concentration is 0.6uM respectively, put it in the environment of 25 ℃, and react for about 12 hours;
[0056] (3) Figure 7 yes image 3 The gel electrophoresis figure of the corresponding product (partial reaction figure of the threshold function module), swimming lane 4 is the reaction lane of M1TQ and Thre, swimming lane 6 Thre* chain, swimming la...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com