Method for constructing target protein interaction network based on high-throughput sequencing
A target protein, high-throughput technology, applied in the field of target protein interaction network construction based on high-throughput sequencing, can solve the problems of limited, labor-intensive, time-consuming and labor-intensive detection capabilities.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1. Screen library conversion process
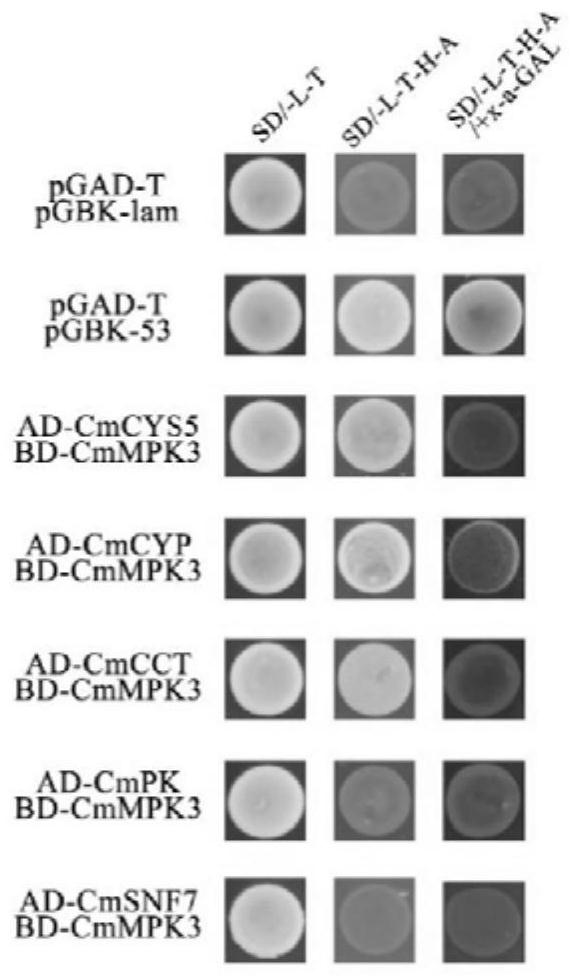
[0044]In this example, the species of the target protein is chrysanthemum. Chrysanthemum CmMPK3 kinase as the target protein, CmMPK3 kinase gene (genbank: MG334203). The known target protein was constructed on the pGBKT7 vector by conventional methods to obtain the BD-bait pGBKT7-CmMPK3, and the yeast AD library was used as the prey library; pGBKT7-CmMPK3 (5 μg) was co-transformed with the yeast AD library (10 μg) using Matchmaker TM These plasmids were co-transformed into the Y2H yeast strain using the Gold Yeast Two-Hybrid System (Clontech).
Embodiment 2 4
[0045] Example 2. Screening of interacting proteins in four-deficiency medium
[0046] Transfer the transformed yeast liquid to 3mL YPDplus (Huayueyang PPT12) medium, culture at 30°C for 4h, centrifuge at 2000rpm for 5min, discard the supernatant, add 1mL sterile water to suspend, and obtain the bacterial liquid; take the obtained 100 μL of bacterial solution was added to 10 mL of four-deficient medium SD / -Leu / -Trp / -His / -Ade liquid medium (Huayueyang *TP0239) at 30°C and 200 rpm for 24h, 72h, and 80h, respectively, and samples were taken.
Embodiment 3
[0047] Embodiment 3.PCR amplification
[0048] The bacterial liquid sampled at 24h, 72h, and 80h was used as a template, and AD-F (SEQ ID NO.1) and AD-R (SEQ ID NO.2) were used as primers to carry out MightyAmp (TaKaRa) PCR amplification according to the procedure shown below. Each sample had 5 repetitions, and the PCR products of the bacterial liquid samples in 24h, 72h, and 80h were obtained respectively.
[0049] PCR program:
[0050]
[0051] The PCR system is:
[0052]
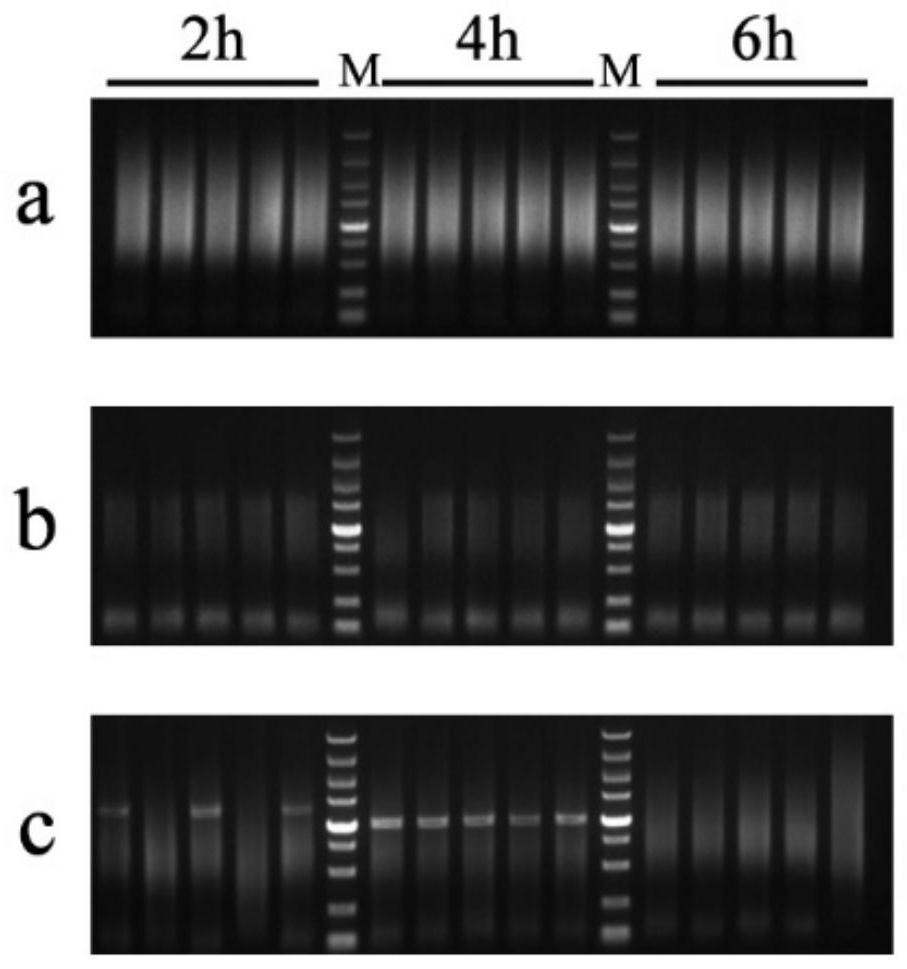
[0053] Agarose gel electrophoresis was used to detect the enrichment of the target protein in the PCR products of the bacterial liquid samples in the three time periods ( figure 1 ), figure 1 It shows that, taking the 5000bp marker as a reference, it is found that the bands are scattered in the range of 500bp-5000bp at 24h and there is no obvious single band, and the length of YPD plus culture has no obvious impact on the results; while at 72h, the PCR amplification product narrows the range to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com