A nucleic acid extraction method and kit
A technology for nucleic acid extraction reagents and extraction methods, applied in the field of nucleic acid extraction methods and kits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
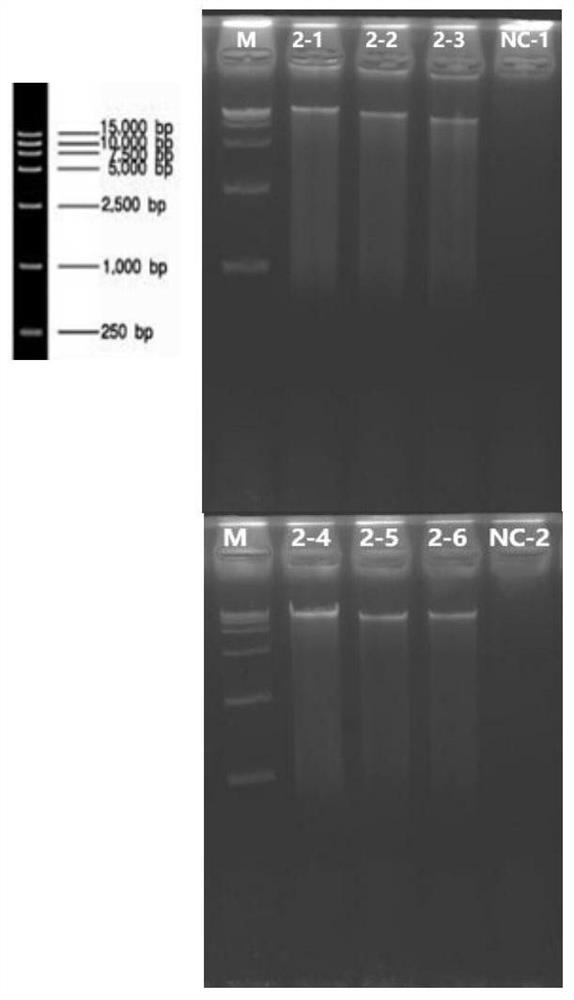
[0119] In order to observe whether different DNA extraction methods affect the results in the field of high-throughput sequencing, two DNA extraction methods were selected, one is a DNA extraction method based on environmental samples published by Gordon et al., as a representative of traditional extraction methods The other method is the fecal DNA extraction kit (product number: D4015) produced by Omega Bio-tek, which is a representative method of DNA extraction from silicon matrix spin columns (abbreviation: Omega kit). Reasons for selection: 1) The traditional extraction method has been used for many years, and it is still recognized, which means that the results are closer to real-time data and are widely accepted. 2) The Omega kit has the main process of the traditional spin column DNA extraction method, which can provide a good reference for subsequent verification of the efficacy of the ingredients of the present invention. Therefore, two extraction methods were used to...
Embodiment 2
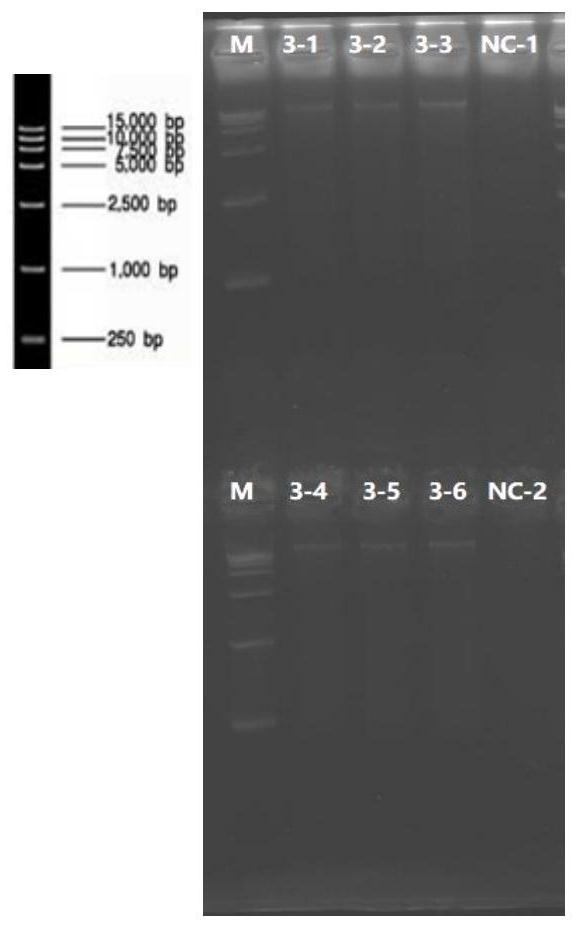
[0126] In order to determine whether each chemical component or its mixture of this method plays the role it should play in the DNA extraction process. The Omega kit with a similar extraction process is replaced with the reagents or components of each extraction process of this method to compare the extraction effect, so as to determine whether the reagents in this method have volatilized their due effect in the link.
[0127] Preparation of reagents for this method:
[0128] 1) Nucleic acid chemical lysis solution: 4M guanidine isothiocyanate solution; 10% w / v N-lauroyl sarcosine sodium salt solution; 5% (w / v) N - Sodium lauroyl sarcosine salt solution.
[0129] 2) Solutions related to impurity removal: 150mM ammonium acetate solution; 120mM ammonium aluminum sulfate dodecahydrate solution (containing 30% v / v PVPP); 20mg / mL proteinase K room temperature storage solution: 25mM Tris-base, 10mM calcium chloride; 200mM Sodium chloride, 50% v / v glycerin, 1% v / v Tween 20, adjuste...
Embodiment 3
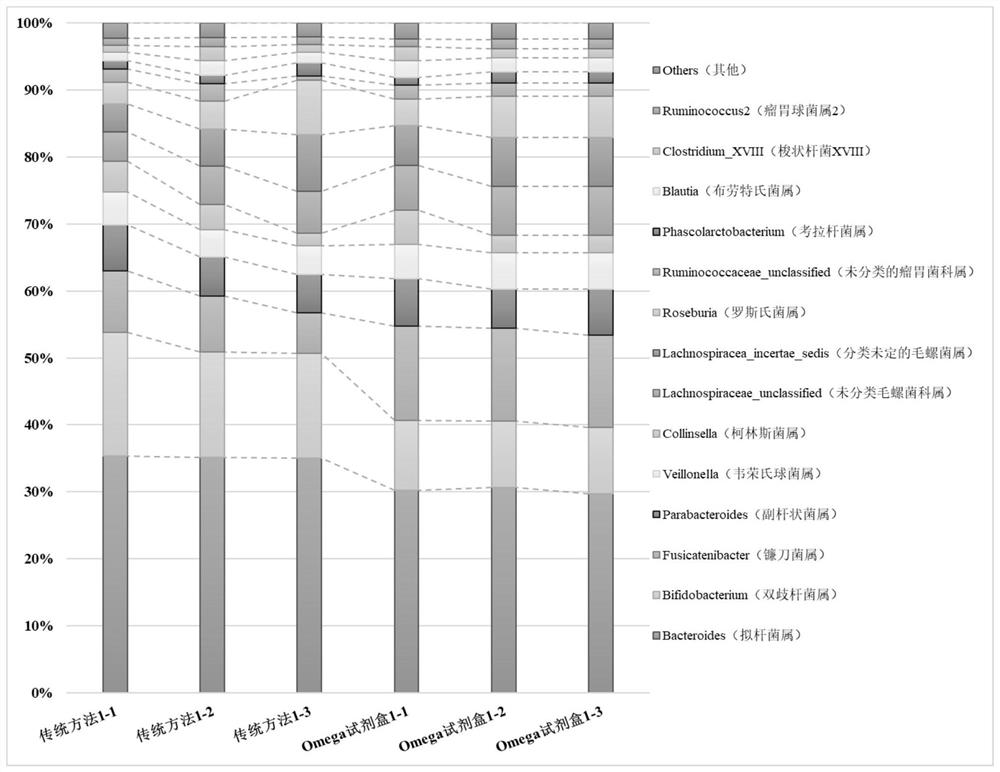
[0177] In order to observe whether this method can more comprehensively obtain the DNA information of various species of samples, the traditional method and this method were used to compare the two sets of DNA extraction processes, and to observe whether there were differences in the final sequencing and bioinformatics analysis results.
[0178] The stool sample of volunteer 11 was selected as the target sample of this test, and the traditional method and the method of the present invention were used to extract the stool DNA respectively, and each method extracted 3 replicates. According to the 16s rRNA gene library construction method officially provided by Illumina (https: / / support.illumina.com.cn / content / dam / illumina-marketing / documents / products / other / 16s-metagenomics-faq-1270-2014-003 .pdf) Complete the library construction of the Miseq sequencing platform and complete the sequencing.
[0179] Bioinformatics analysis was performed as follows.
[0180] The raw data (Rawdat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com