Chitin deacetylase gene, chitin deacetylase and preparation method and application of chitin deacetylase
A deacetylase and chitin technology, applied in the field of genetic engineering, can solve problems such as low fermentation activity, and achieve the effects of high-efficiency expression, good stability and lower production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0044] Correspondingly, the embodiment of the present invention also provides a preparation method of chitin deacetylase CdaBa, which comprises the following steps:
[0045] S1. Provide chitin deacetylase gene cdaba, expression vector and expression strain;
[0046] S2. Amplifying the chitin deacetylase gene cdaba, and connecting the obtained amplified product with an expression vector to obtain a recombinant expression vector;
[0047] S3. Transforming the recombinant expression vector into an expression strain to obtain a recombinant expression strain;
[0048] S4. Cultivate the recombinant expression strain to obtain chitin deacetylase CdaBa;
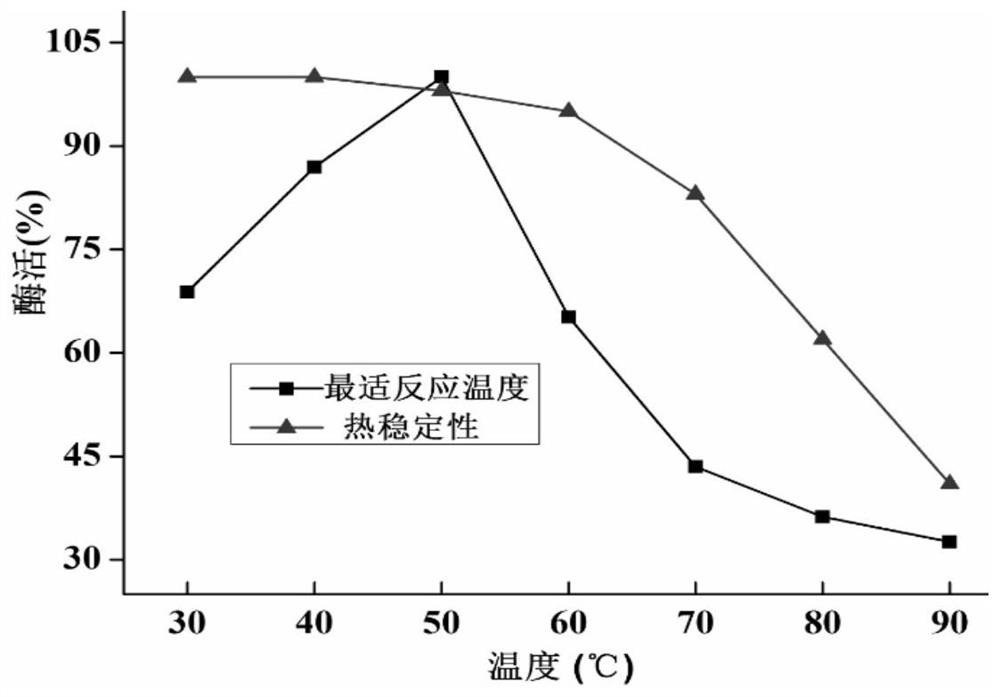
[0049] Wherein, the nucleotide sequence of the chitin deacetylase gene cdaba is the nucleotide sequence shown in SEQ ID NO: 1, or the nucleotide sequence shown in SEQ ID NO: 1 through deletion, insertion or replacement The resulting nucleotide sequence has the same function; the amino acid sequence of chitin deacetylase CdaBa is th...
Embodiment 1
[0075] This embodiment provides chitin deacetylase gene sequence analysis and codon optimization process, including the following steps:
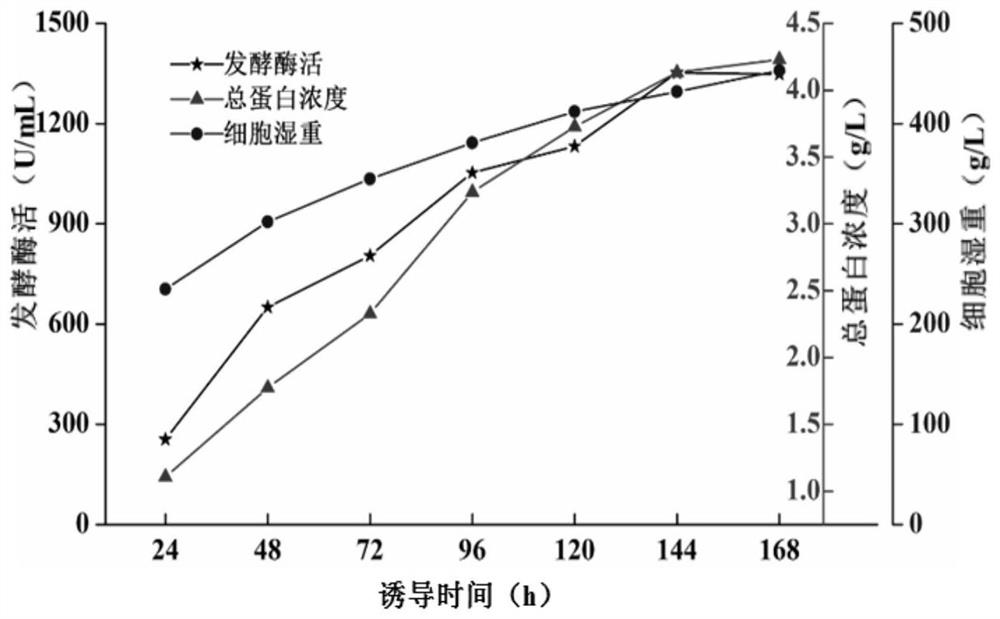
[0076] (11) Based on the many shortcomings of the natural bacteria Bacillus atrophaeus, it is proposed to improve its expression efficiency through heterologous recombination expression. Among them, the Pichia pastoris expression system was selected as the heterologous expression host. The first step of heterologous recombination expression is to find the Bacillus atrophaeus chitin deacetylase gene. Its whole genome sequence was submitted to the NCBI database in 2017, and its accession number is CP024051.1. By analyzing its genome data, it was found that the nucleotides in the interval 4031023-4031853 of its genome are the coding gene of chitin deacetylase CdaBa. The full length of the gene is 831bp, encoding 276 amino acids. The first 38 amino acids of chitin deacetylase CdaBa were found to be its signal peptide through the prediction a...
Embodiment 2
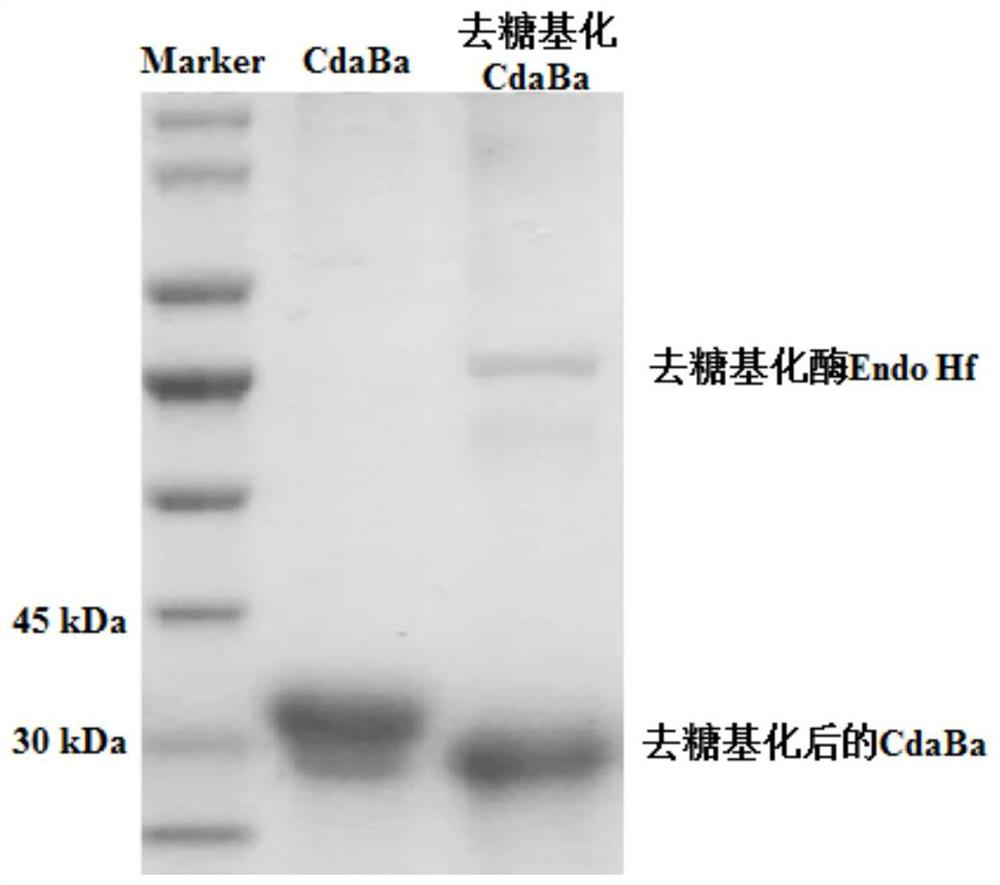
[0079] This example provides a construction process of a recombinant expression vector containing the chitin deacetylase gene cdaba obtained in Example 1. The recombinant expression vector uses pPICZαA as the expression vector, and the signal peptide used is the α signal peptide carried by pPICZαA. Therefore, the signal peptide of cdaba itself needs to be removed during the construction process. details as follows:
[0080] (13) Design a pair of primers cdaba-F and cdaba-R according to the sequence of the synthetic gene cdaba, its nucleotide sequence is shown in SEQ ID NO:3-4 respectively, for amplifying the cdaba gene of removing signal peptide;
[0081] (14) Obtain the gene cdabas that removes the signal peptide by PCR amplification, digest the expression vector pPICZαA and the gene cdabas with restriction endonucleases EcoRI and XbaI respectively overnight, and purify and recover the expression vector pPICZαA and cdabas overnight. ligation reaction;
[0082] (15) Transfer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com