Application of protein encoded by salmonella phage gene as gram-negative bacterium lyase
A technology for Gram-negative bacteria and Salmonella, applied in the direction of lyase, application, genetic engineering, etc., can solve the problems of few application examples and the inability to produce Gram-negative bacteria decidual cells, etc., and achieve the effect of high lysis effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
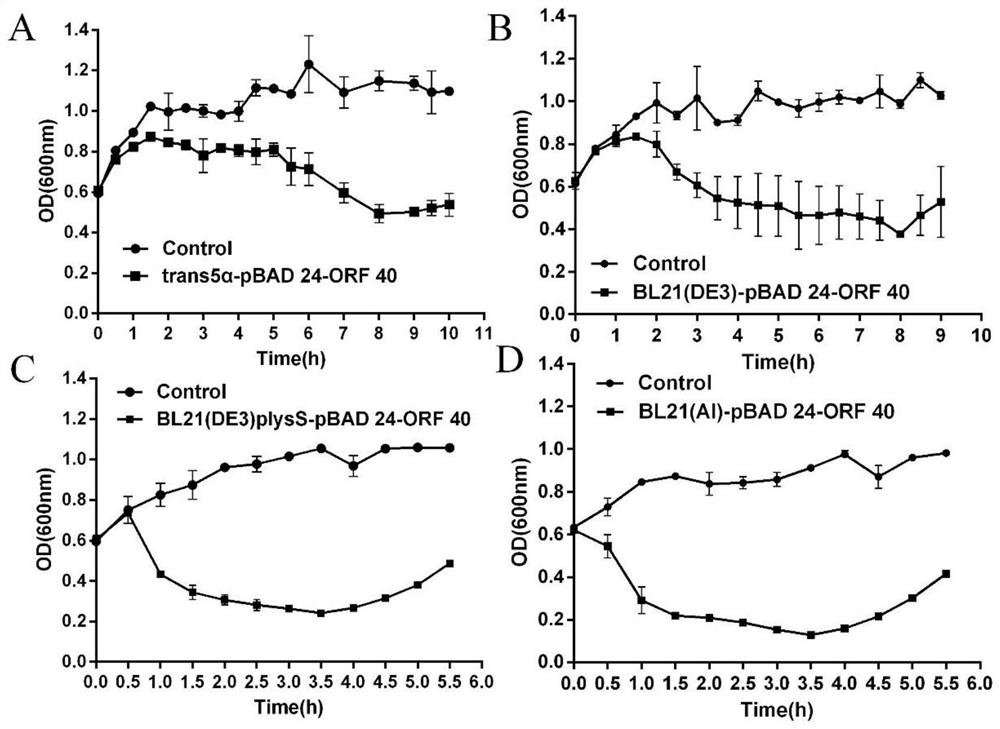
[0020] Construction of Engineering Strain and Determination of Self-lysis Curve
[0021] 1. Lyase gene cloning and pBAD24-ORF 40 vector construction
[0022] Using the base sequence of ORF 40 (shown in SEQ ID NO.1, the corresponding amino acid sequence is shown in SEQ ID NO.2) as a template, the upstream and downstream primers ORF 40-F and ORF 40-R for inserting the gene were designed, and the plasmid The sequence of pBAD 24 was used as a template to design the upstream and downstream primers pBAD24-F and pBAD24-R for plasmid linearization, and the primers ORF 40-F and ORF 40-R for inserting the gene had the same The source sequence was synthesized by Shanghai Shenggong Company.
[0023] orf 40-f: ttgggctagcaggaggaattcaccatgaagtcgcgtcgaa ga aga
[0024] orf 40-r:caaaacagccaagcttgcacgttattattgcaccggttttgaca
[0025] pbad24-f:taacgtgcaagcttggctgttttg
[0026] pbad24-r:ggtgaattcctcctgctagcccaa
[0027] The procedure for PCR amplification of the target gene ORF40 was as follo...
Embodiment 2
[0036] Escherichia coli decidual cells
[0037] 1. Identification of chloramphenicol-resistant pBAD 24-ORF 40 recombinant plasmid
[0038] Make Escherichia coli X7213 competent cells, and transform the constructed recombinant plasmid pBAD24-ORF 40 into X7213 competent cells.
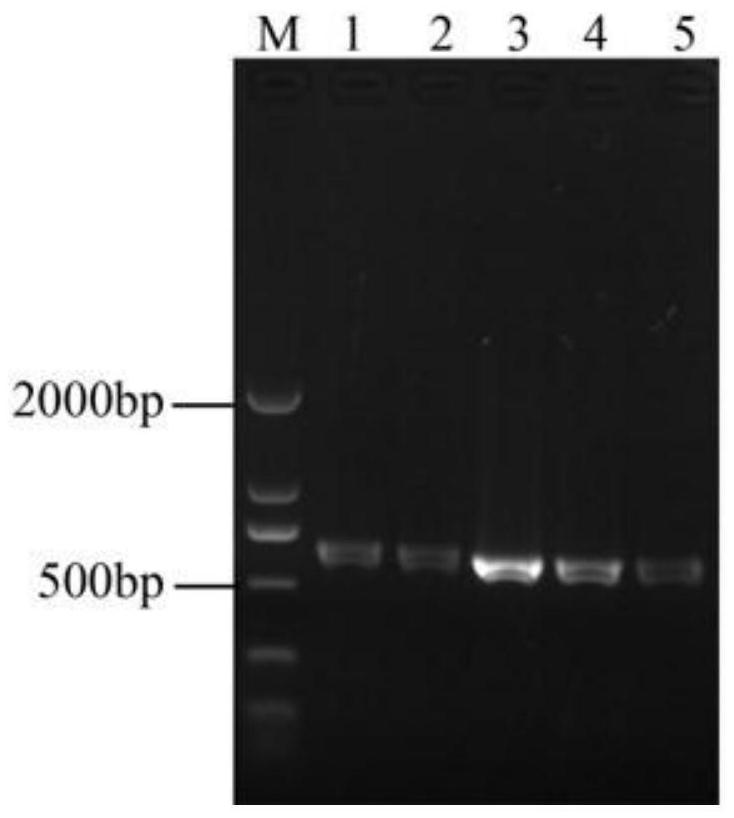
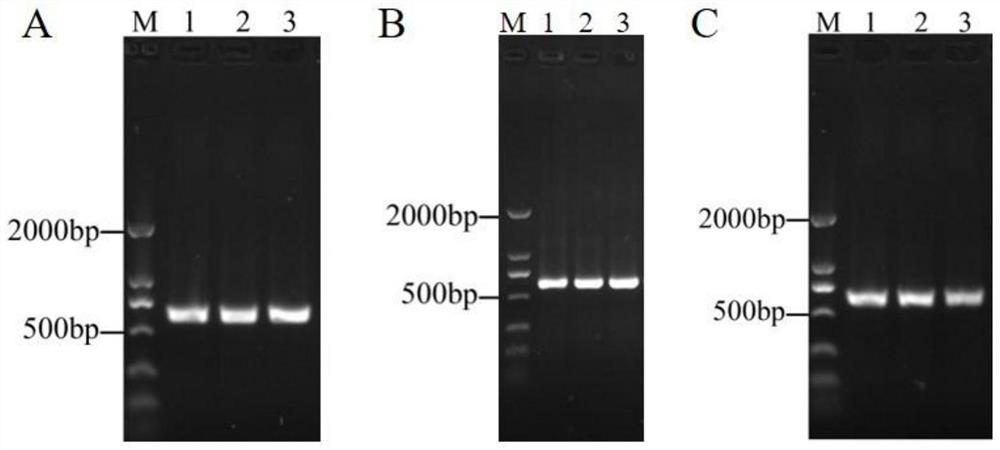
[0039] Use ORF 40-F and ORF 40-R as primers to verify the transformants by PCR, and take the PCR products for gel electrophoresis, and the bands shown are consistent in size with the target bands, as shown in Figure 4 Shown in A.
[0040] 2. Conjugative transfer to Salmonella typhimurium and pathogenic E. coli
[0041] Salmonella typhimurium ATCC 13311 and Escherichia coli ATCC 700728 were used as recipient bacteria, and Escherichia coli X7213 carrying the recombinant plasmid pBAD 24-ORF 40 resistant to chloramphenicol was used as donor bacteria to construct a decidual strain by conjugative transfer. Pick the transformants on the transformation plate to inoculate and culture, use the bacterial soluti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com