Self-induced protein expression vector based on AI-2 quorum sensing and application
A quorum sensing and AI-2 technology, applied in the field of molecular biology, can solve the problems of inhibiting cell viability, high expression of exogenous genes, and affecting the viability of host strains, and achieve the effect of less inclusion bodies and high protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
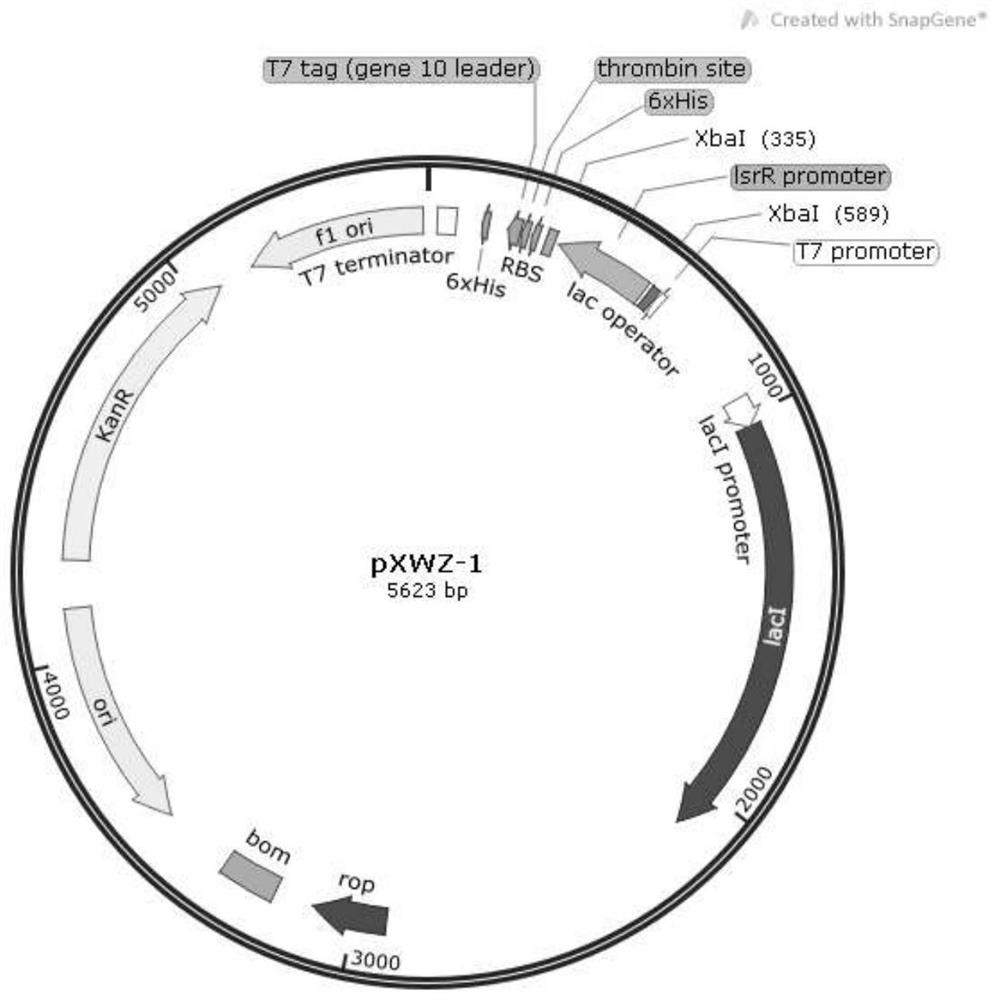
[0032] Example 1 Construction of vector pXWZ-1
[0033] The primers were designed according to the sequence of the Escherichia coli lsrR promoter gene reported on NCBI, and the genome of the Escherichia coli model strain MG1655 with the lsr operon was used as a template. Use plsrR-Xba I-f, plsrR-Xba I-r: as primers to amplify the lsrR promoter sequence.
[0034] Upstream primer: plsrR-Xba I-f:5'-GC TCTAGA AATTCATTCTTCACTTTGAA-3' (the underline is the restriction site of XbaI, SEQ ID NO.3)
[0035] Downstream primer: plsrR-Xba I-r:5'-GC TCTAGA ATTTCCCCCGTTCAGTTTTG-3' (the underline is the restriction site of XbaI, SEQ ID NO.4)
[0036] The amplified lsrR promoter fragment and the pET28(a)+ plasmid were respectively digested with restriction endonuclease Xba I, incubated at 37°C for 2 hours for agarose gel electrophoresis and gel recovery, and the recovered DNA fragment was named as A1, A2. Mix A1 and A2 in a certain system, connect overnight with DNA ligase, and then tra...
Embodiment 2
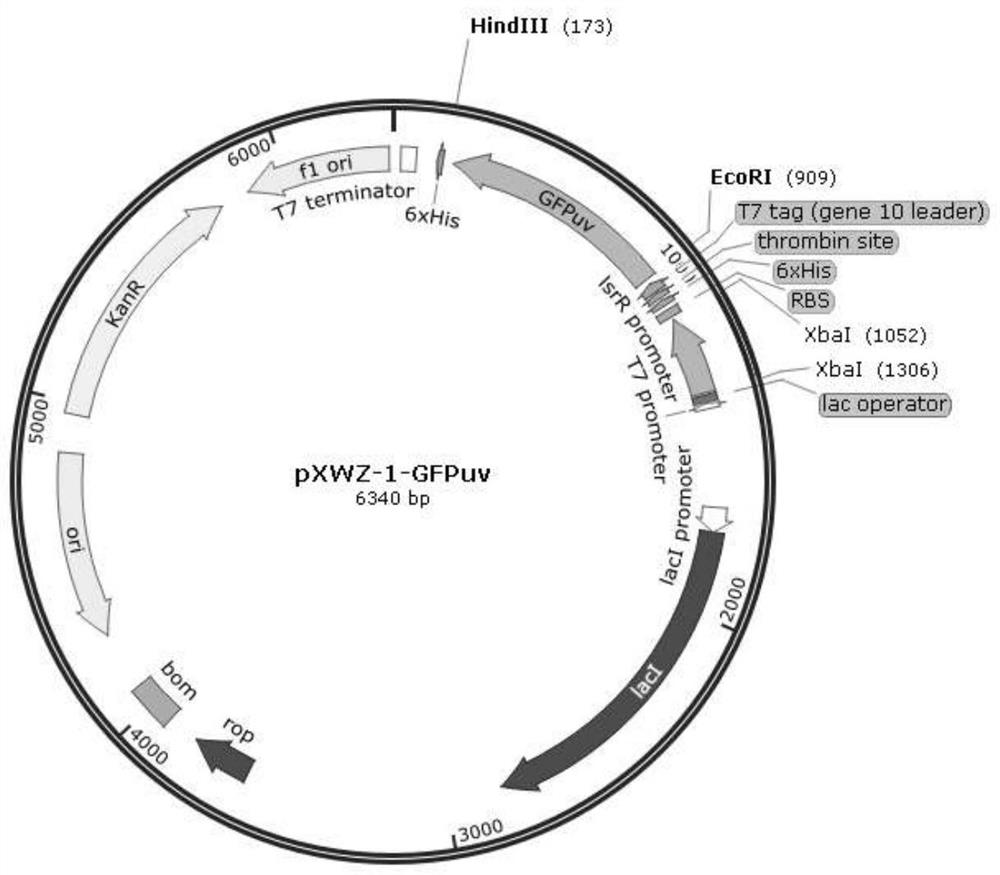
[0037] Example 2 Construction of vectors pET28a-GFPuv and pXWZ-1-GFPuv
[0038] Extract the pGFPuv plasmid, use the pGFPuv plasmid as a template, gfp-EcoR I-f, gfp-Hind III-r as primers, PCR amplify the GFPuv fragment, connect it to the vector pET28(a)+ and pXWZ-1 respectively, and construct the expression vector pET28a- GFPuv and pXWZ-1-GFPuv.
[0039] Upstream primer: gfp-EcoR I-f:5'-CCG GAATTC ATGAGTAAAGGAGAAGAACT-3' (underlined is the restriction site of EcoR I)
[0040] Downstream primer: gfp-Hind III-r:5'-CCC AAGCTT TTATTTGTATAGTTCATCCA-3' (the restriction site of Hind III is underlined)
[0041] The amplified GFPuv fragment, pET28(a)+ plasmid, and pXWZ-1 plasmid were respectively digested with restriction endonucleases EcoR I and Hind III, incubated at 37°C for 2 hours for agarose gel electrophoresis and gel recovery. The recovered DNA fragments were named B1, B2, and B3, respectively. B1 and B2, B1 and B3 were mixed in a certain system, connected overnight with ...
Embodiment 3
[0042] Embodiment 3 expresses green fluorescent protein
[0043] 1. Construction of engineered bacteria
[0044] The expression vectors pET28a-GFPuv and pXWZ-1-GFPuv were respectively transformed into Escherichia coli BL21(DE3) by chemical transformation, and spread on LB solid plates containing 50 μg / mL kanamycin. Incubate overnight in a 37°C incubator. Single clonal colonies were selected for PCR identification.
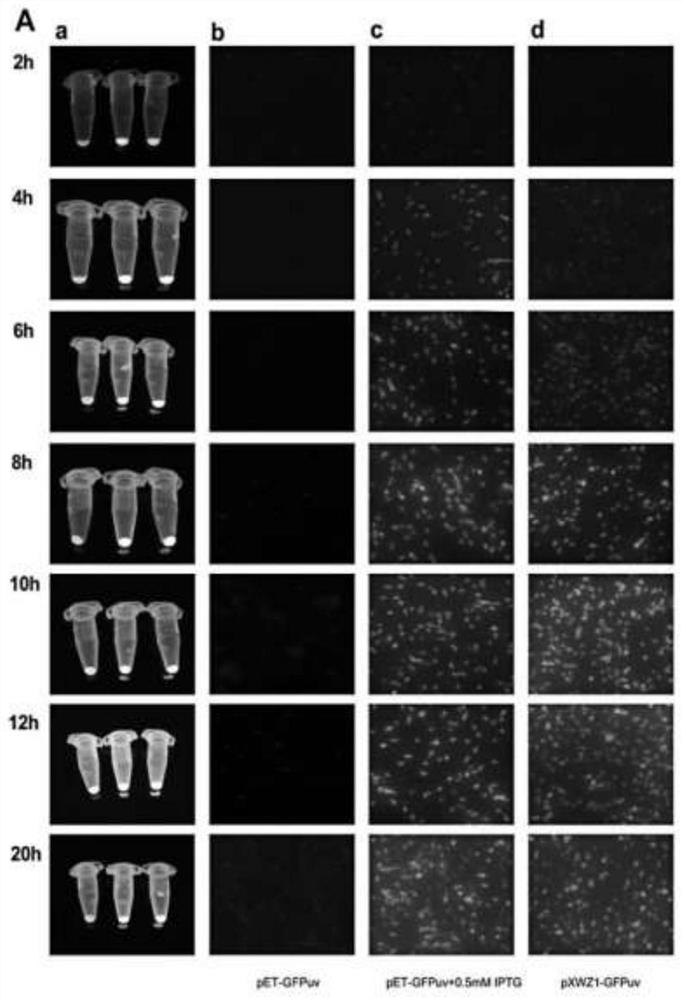
[0045] 2. Using a fluorescence microscope to monitor the fluorescence intensity of GFPuv
[0046] The BL21(DE3) bacterial solution containing the expression vectors pET-GFPuv and pXWZ-1-GFPuv was transferred to 100 mL fresh LB medium containing 50 μg / mL kanamycin at a ratio of 1:100 (v / v). Marked as ①, ②, ③ respectively. Among them, ① sample is BL21 / pET28-GFPuv, ② sample is BL21 / pET28-GFPuv induced by adding final concentration of 0.5mM IPTG, and ③ sample is BL21 / pXWZ-1-GFPuv. Cultivate at 37°C with shaking at 150rpm until OD600=0.3, add 0.5mM IPTG to ②, and p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com