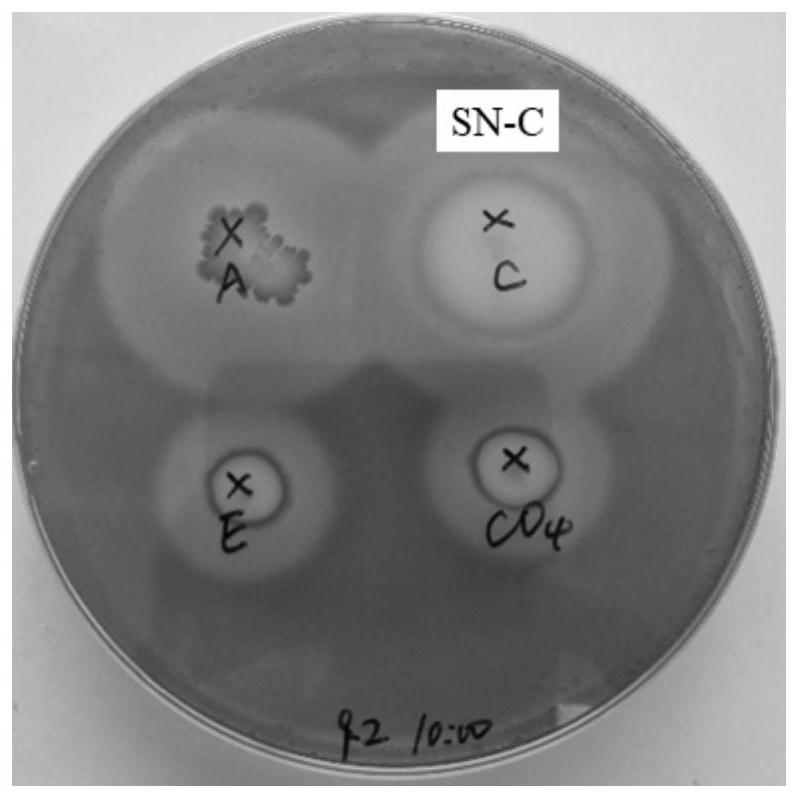
Screening and application of buffalo rumen-derived bacillus amyloliquefaciens
A technology for dissolving starch spores and water buffalo, applied in the field of agricultural microorganism application, can solve problems such as insufficient cellulose degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Isolation and identification of bacterial strains
[0038] 1. The preparation of culture medium:
[0039] Table 1 Buffer A
[0040]
[0041] NOTE: This buffer is used to prepare enrichment media.
[0042] Table 2 Buffer B
[0043]
[0044] NOTE: This buffer is used to prepare enrichment media.
[0045] Table 3 enrichment medium
[0046]
[0047]
[0048] Note: Resazurin is blue, and the color of the medium will turn pink after sterilization, and the solution is colorless when the medium does not contain oxygen.
[0049] Prepare the antifungal agent according to the formula in Table 4. After the preparation is completed, use a 0.45 μm filter to filter and sterilize, aliquot into 1.0 mL, and store at 4°C.
[0050] The configuration of table 4 antifungal agent
[0051]
[0052] Note: Cycloheximide is a highly toxic drug that inhibits yeast, mold and protozoa; the solvent used for nystatin is DMSO, dissolve a small amount of DMSO and then mix...
Embodiment 2
[0109] Example 2: Safety Evaluation of Bacillus amyloliquefaciens SN-C strain
[0110] 1. Virulence gene amplification (1) Identification of virulence gene positive strains
[0111] The gene primers of non-hemolytic enterotoxin NHE (nheA, nheB, nheC), hemolysin Hbl (hblA, hblC, hblD), cytotoxin CytK, enterotoxin T (BceT) and enterotoxin FM (EntFM) were used to determine the virulence Amplify the gene-positive Bacillus cereus, detect the virulence genes contained in it, and use 1% agarose gel electrophoresis (120V, 90mA), and the positive identification of the virulence gene is that the given Bacillus cereus contains nheA, nheB, and nheC , EntFM, hblA, hblC, hblD, CytK, BceT a total of 9 virulence genes; gene primer sequences are shown in Table 11, amplification results are shown in Figure 7 .
[0112] Table 11 Virulence gene PCR primer sequence
[0113]
[0114] (2) Amplification of virulence genes of Bacillus amyloliquefaciens SN-C strain
[0115] Bacterial Genomic DN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com