Method for producing ice nucleation protein by fermentation of recombinant Escherichia coli
A technology for recombining Escherichia coli and ice nucleation protein, which is applied in the fields of genetic engineering and microbial fermentation, can solve the problems of low activity, difficulty in stable production, and high expression of ice nucleation active protein, and achieves high activity, short lag period, and moderate expression. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
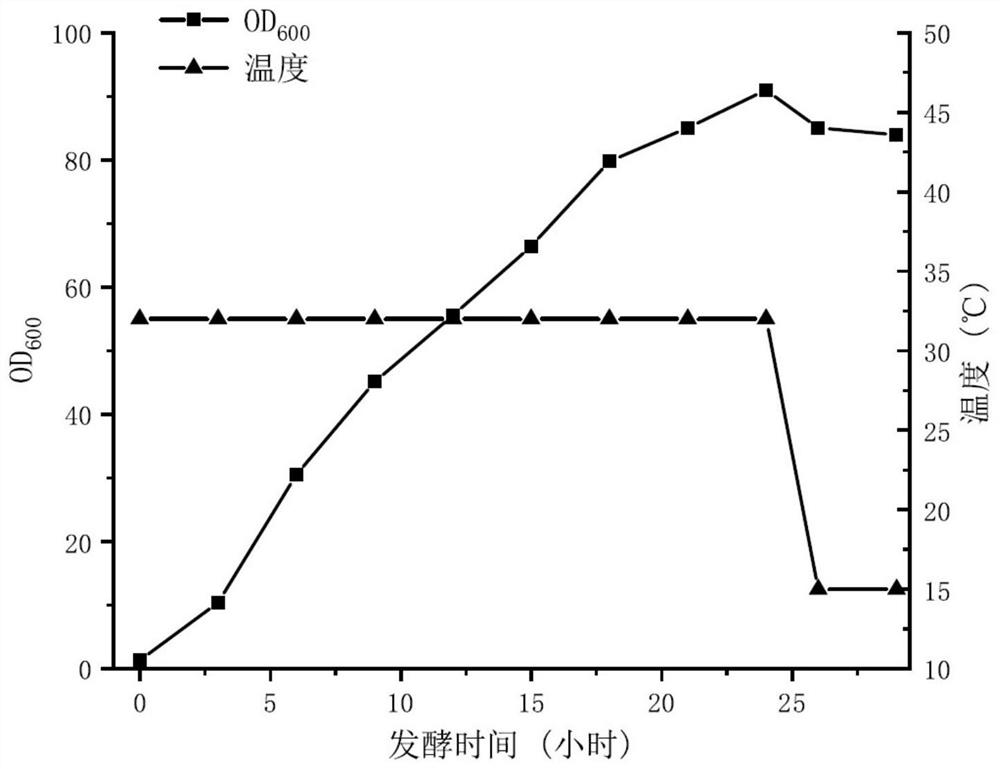
[0060] Embodiment 1 Utilizes the method for recombinant escherichia coli fermentation to produce ice nuclei protein
[0061] 1. Construction of recombinant Escherichia coli TDTC-inp001 producing ice nucleation protein
[0062] 1.1 Amplification of ice nucleoprotein gene inp
[0063] Using the genomic DNA of Pseudomonas syringae (Psudomonas syringae) TDTC-IN 13 as a template, PCR amplification was performed with the following primers:
[0064] inp-F: ctaacaggaggaattaaccatgaatctcgacaaggcgttg
[0065] inp-R: gagctcggatccccatcgatctactcgacctctatccagtc
[0066] The high-efficiency fidelity enzyme Phanta Max Super-Fidelity DNA Polymerase from Vazyme Biotech Co., Ltd. was used for PCR amplification. The PCR amplification program was: 95°C for 3min; 30 cycles of 95°C for 15s, 58°C for 15s, and 72°C for 2min; 72°C for 5min. The obtained PCR product was subjected to electrophoresis and then gel-cut, and the gel recovery kit of Omega Company was used for gel recovery (operated accordi...
Embodiment 2
[0111] Embodiment 2 Utilizes the method for recombinant escherichia coli to ferment and produce ice nuclei protein
[0112] 1. The recombinant Escherichia coli TDTC-inp004 producing ice nucleation protein, its construction method is as follows:
[0113] 1.1 Amplification of ice nucleoprotein gene inp
[0114] Using the genomic DNA of Pseudomonas syringae (Psudomonas syringae) TDTC-IN 13 as a template, PCR amplification was performed with the following primers:
[0115] inp-F: ctttaagaaggagatataccatgaatctcgacaaggcgttg
[0116] inp-R: gcaagcttgtcgacctgcagctactcgacctctatccagtc
[0117] The high-efficiency fidelity enzyme Phanta Max Super-Fidelity DNA Polymerase from Vazyme Biotech Co., Ltd. was used for PCR amplification. The PCR amplification program was: 95°C for 3min; 30 cycles of 95°C for 15s, 58°C for 15s, and 72°C for 2min; 72°C for 5min. The obtained PCR product was subjected to electrophoresis and then gel-cut, and the gel recovery kit of Omega Company was used for ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com