Method for determining the response of a malignant disease to an immunotherapy
A technology for immunotherapy and disease, applied in biochemical equipment and methods, pharmaceutical formulations, antibody medical components, etc., can solve different problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
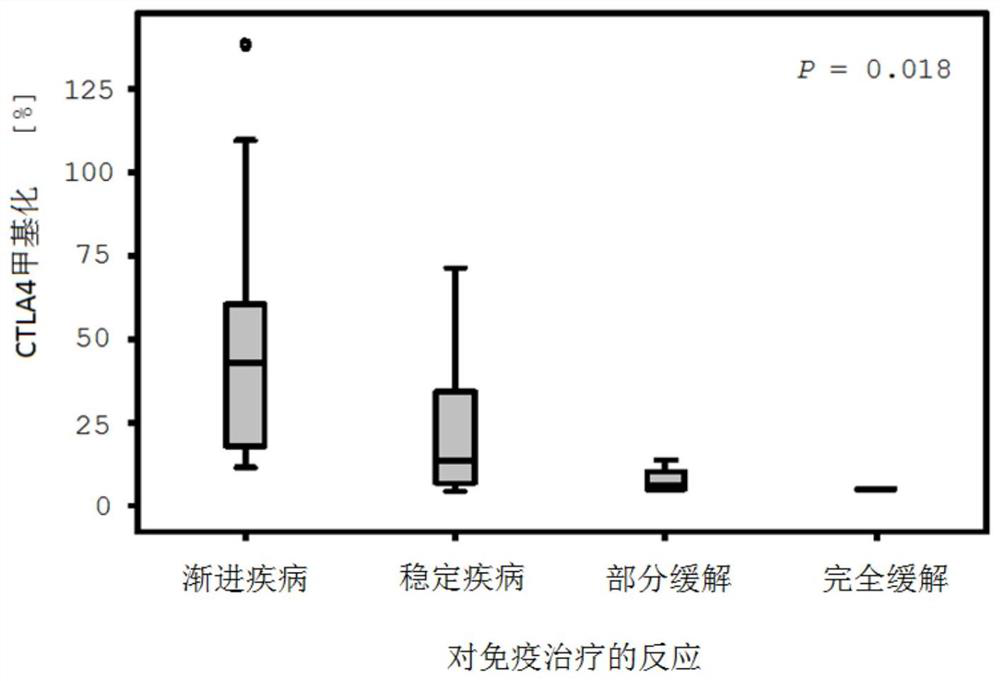
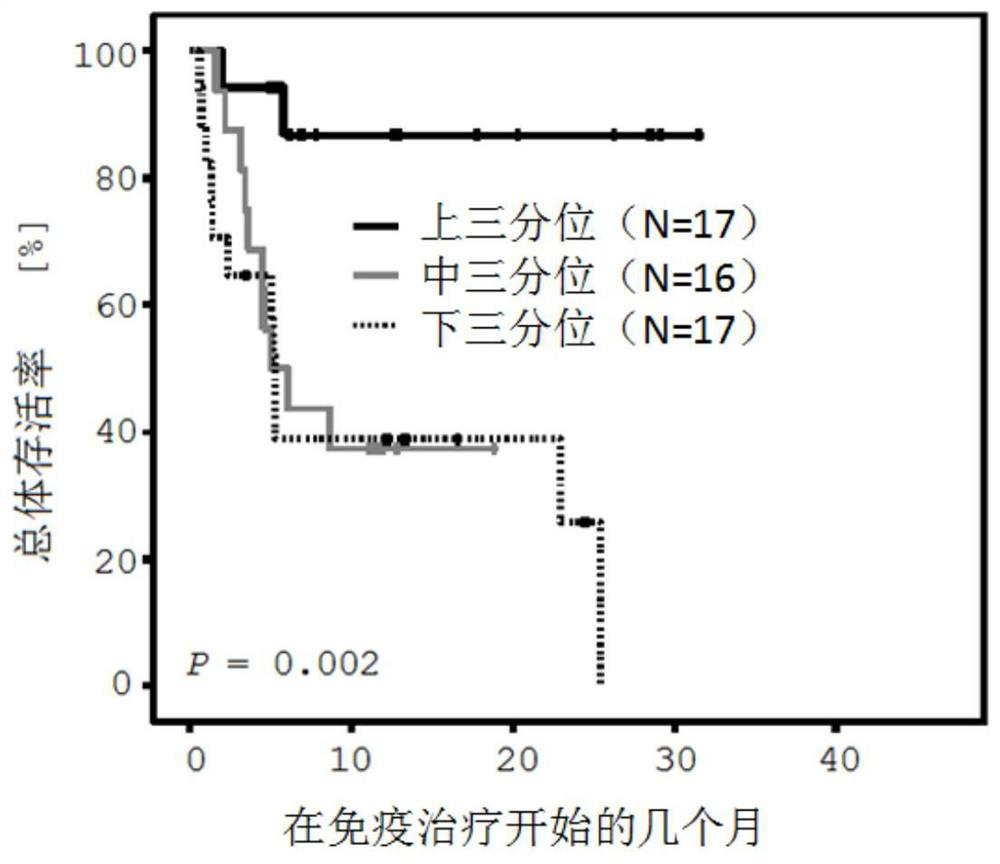
[0095] Example 1: CTLA4-based DNA methylation predicts malignant disease to inhibit PD-1 immune checkpoint signaling pathway Clinical Studies of Responses to
[0096] The patient cohort examined included a total of 50 patients diagnosed with metastatic malignant melanoma. Before immunotherapy, tumor tissue samples were removed from patients, fixed with formalin, and embedded in paraffin. Patients were treated with anti-PD-1 immune checkpoint blockade with pembrolizumab or nivolumab between October 2014 and April 2017.
[0097] For DNA methylation analysis, thin sections of tumor tissue samples with a thickness of 10 μm were first made, and then mounted on glass carriers. Based on HE sections, tumor areas were identified by pathological evaluation and scraped from glass carriers for further processing. Bisulfite-converted DNA was delivered from the tumor area using the InnuCONVERT Bisulfite All-in-One Kit (Analytik Jena, Jena, Germany) according to the manufacturer's inst...
Embodiment 2
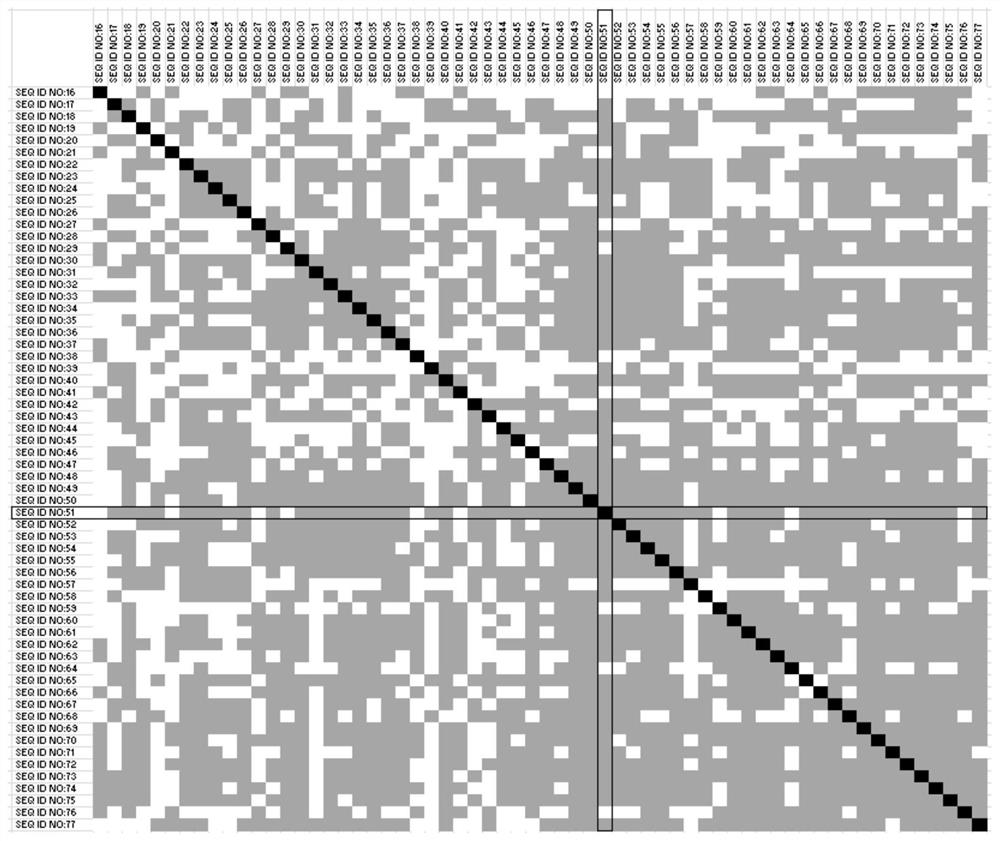
[0105] Example 2: Determination of co-methylation of CTLA4, CD86, CD28, CD80 and ICOS in various malignant diseases
[0106] In the context of the present invention, it is also recognized that a patient's response to immunotherapy can be determined not only by DNA methylation analysis of CTLA4, but also by the immunomodulatory genes whose DNA methylation correlates with the DNA methylation of CTLA4 DNA methylation analysis to determine. The immunoregulatory genes CD28, CD80, CD86 and / or ICOS are particularly suitable here. These genes are functionally related to CTLA4. In addition, CD28 and ICOSim in the genome are also located near CTLA4.
[0107] Co-methylation of CTLA4, CD28, CD80, CD86, and ICOS was examined using genome-wide DNA methylation analysis, such as the Infinium HumanMethylation45 bead chip (Illumina, Inc., San Diego, CA, USA). TCGA Research Network ( http: / / cancergenome.nih.gov / ) describes the generation of HumanMethylation450 BeadChip raw data. In conc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com