Neutralizing antibody for resisting novel coronavirus SARS-Cov-2 and application thereof
A sars-cov-2, coronavirus technology, applied in the direction of antiviral agents, antiviral immunoglobulins, applications, etc., can solve problems such as affinity differences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
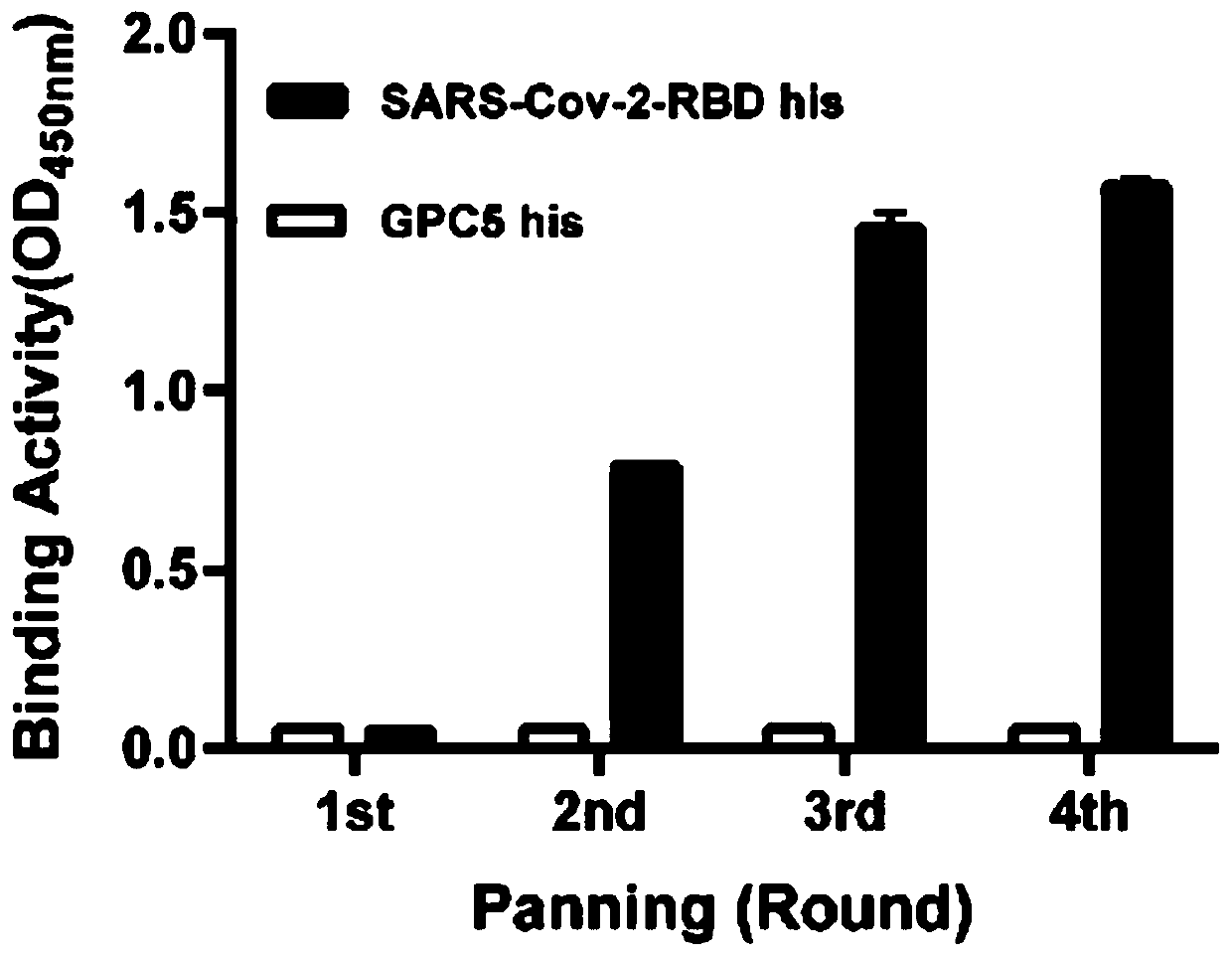
[0037] Example 1. Screening of fully human antibodies targeting SARS-Cov-2-RBD
[0038] Using phage display technology, the SARS-Cov-2-RBD-his protein was used as the positive antigen, and the SARS-Cov-1-RBD-hFc was used as the negative antigen. is 1.47x10 8 ) for differential screening.
[0039] Immunoplates were coated with 50 μg / ml SARS-Cov-2-RBD his antigen and SARS-Cov-1-RBD hFc antigen at 4°C overnight; blocked with PBS solution containing 5% nonfat dry milk and 0.1% Tween-20 at room temperature Immunoplate for 1 hour; 12 pfu was mixed 1:1 with 10% nonfat dry milk PBS solution, incubated at room temperature for 2 hours, added to the blocked SARS-Cov-1-RBD hFc antigen immunoplate (100 μl / well), incubated at room temperature for 1 hour, and pre-adsorbed with negative antigen ; After pre-adsorption, the supernatant was transferred to the SARS-Cov-2-RBD his antigen immune plate (100 μl / well), which was sealed, and incubated at room temperature for 1 hour. The immune plat...
Embodiment 2
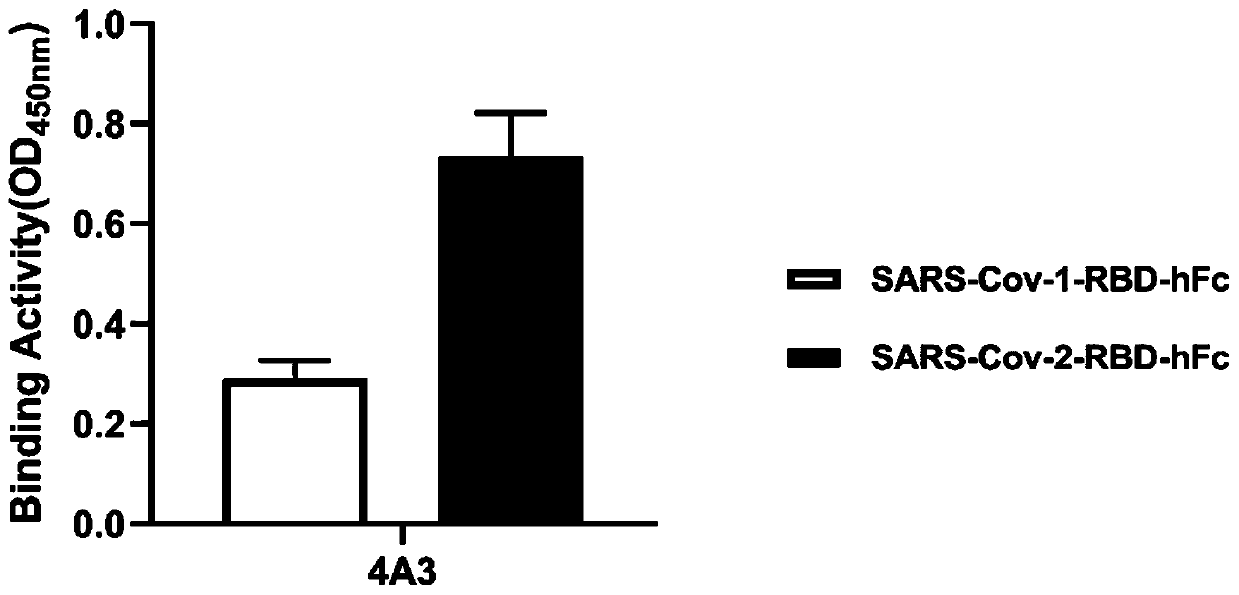
[0050] Example 2. Antigen specificity analysis of antibodies
[0051] In this example, ELISA was used to detect the binding of the 4A3 phage of Example 1 to the SARS-Cov-2-RBD protein.
[0052] The specific process is as follows: use 5 μg / ml SARS-Cov-1-RBD-hFc and SARS-Cov-2-RBD-hFc to coat the immune plate overnight at 4°C; The PBS solution of 0.05% Tween-20 was used to block the immune plate for 1 hour at room temperature; the phage of 4A3 was added to the blocked immune plate (50 μl / well), and incubated at room temperature for 1 hour; the immune plate was washed 3 times with 0.05% Tween-20 in PBS (340 μl / well). ); Mix HRP / Anti-M13 Monoclonal conjugate with PBS solution containing 5% nonfat milk powder and 0.05% Tween-20 at a ratio of 1:4000, add it to the washed immune plate (50μl / well), and incubate at room temperature for 1 hour; Wash the immune plate 5 times with 0.05% Tween-20 in PBS; add TMB color development solution to the immune plate (100 μl / well), and after 3 min...
Embodiment 3
[0054] Example 3. Expression and purification of antibodies
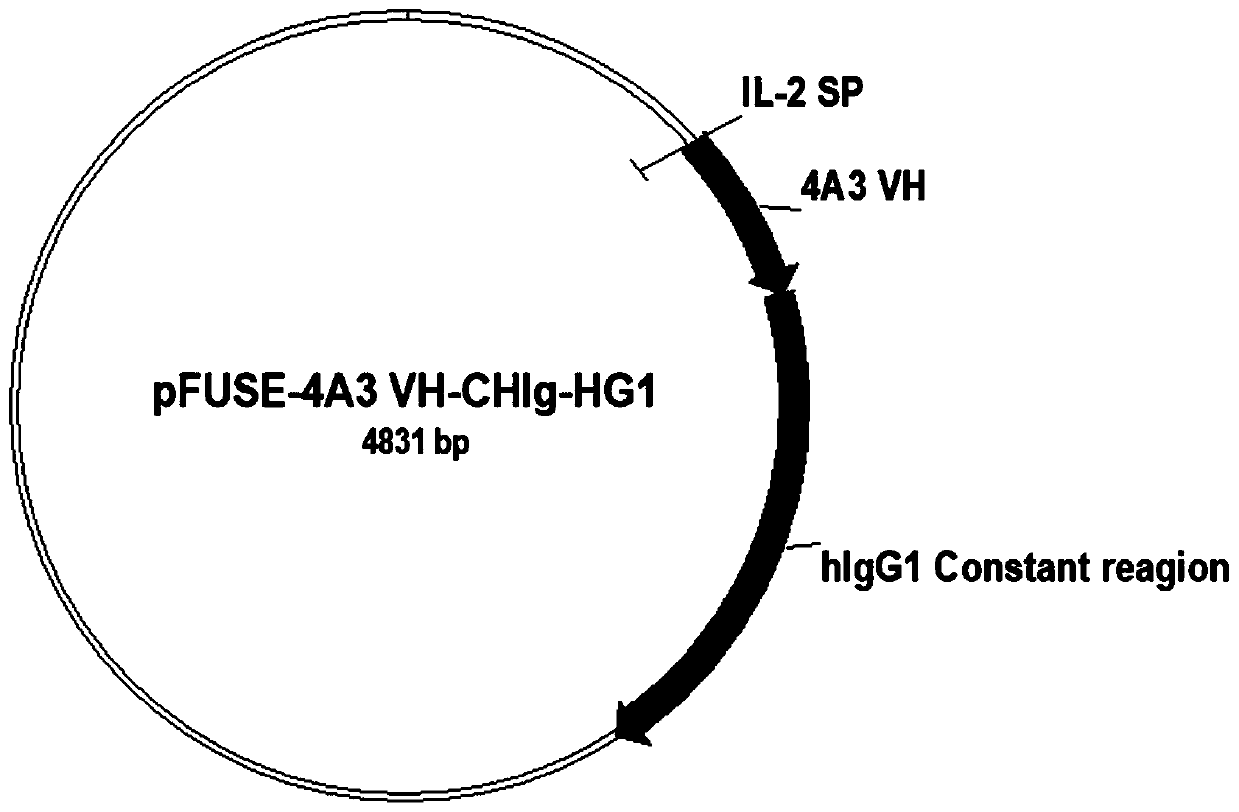
[0055] The heavy chain variable region sequence and light chain variable region sequence of the 4A3 scFv sequence were inserted into the pFUSE-CHIg-HG1 and pFUSE2-CLIg-hk vectors (Invivogen, San Diego, CA), respectively, to construct an adult IgG1 expression plasmid, such as image 3 , Figure 4 shown. The above plasmids were co-transfected in 293T cells, and the supernatant was collected and purified with a protein A-Agarose separation column. The purity of the antibody was detected by SDS-PAGE. Figure 5 shown.
[0056] The specific process is as follows: insert the heavy chain variable region sequence and light chain variable region sequence of the 4A3 scFv sequence into pFUSE-CHIg-HG1 and pFUSE2-CLIg-hk (Invivogen, San Diego, CA), respectively, to construct an adult IgG expression plasmid. 5 million HEK293T cells were seeded in a cell culture dish with DMEM medium supplemented with 10% fetal bovine serum, 100...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com