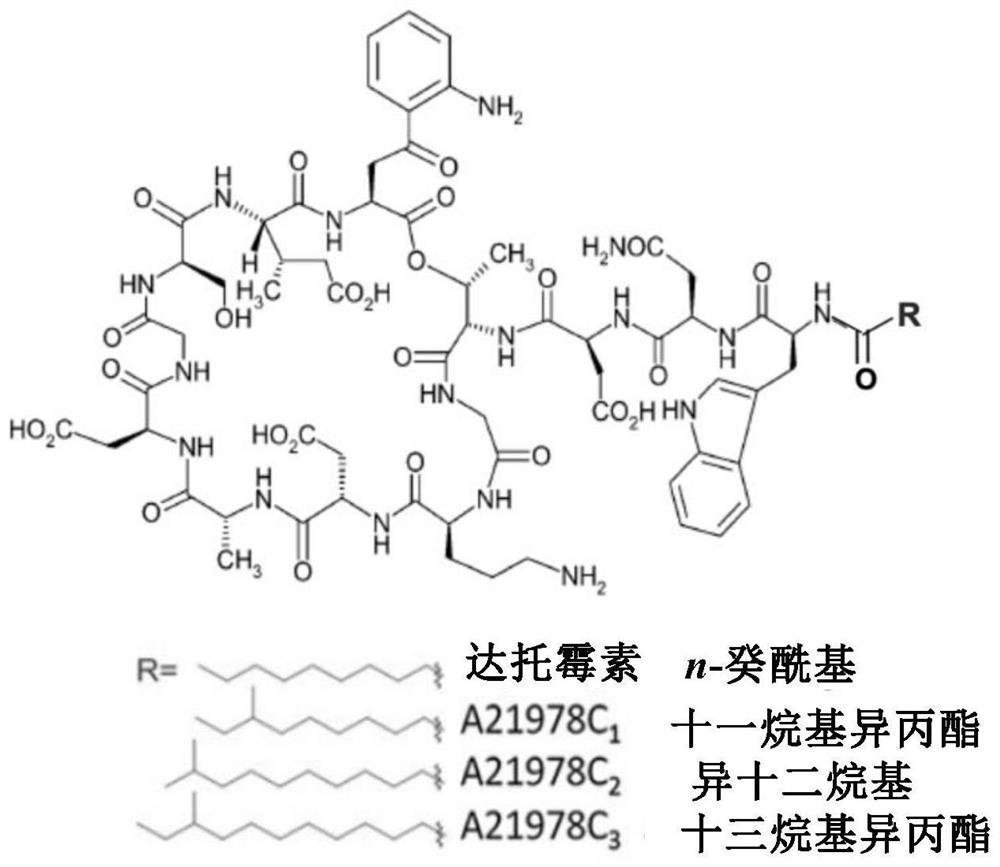
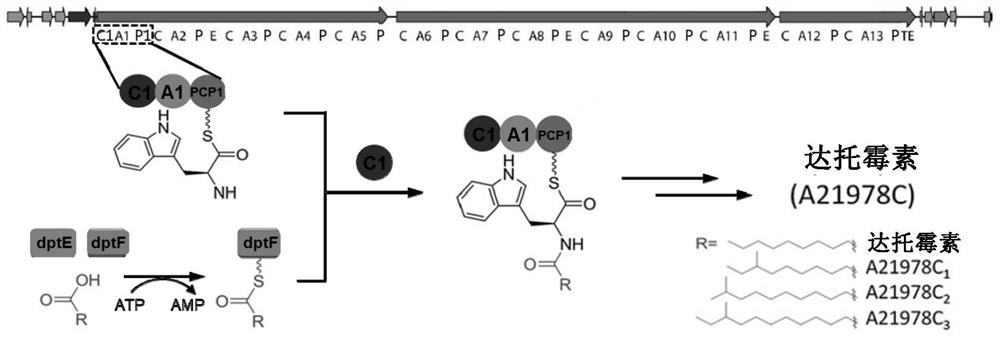
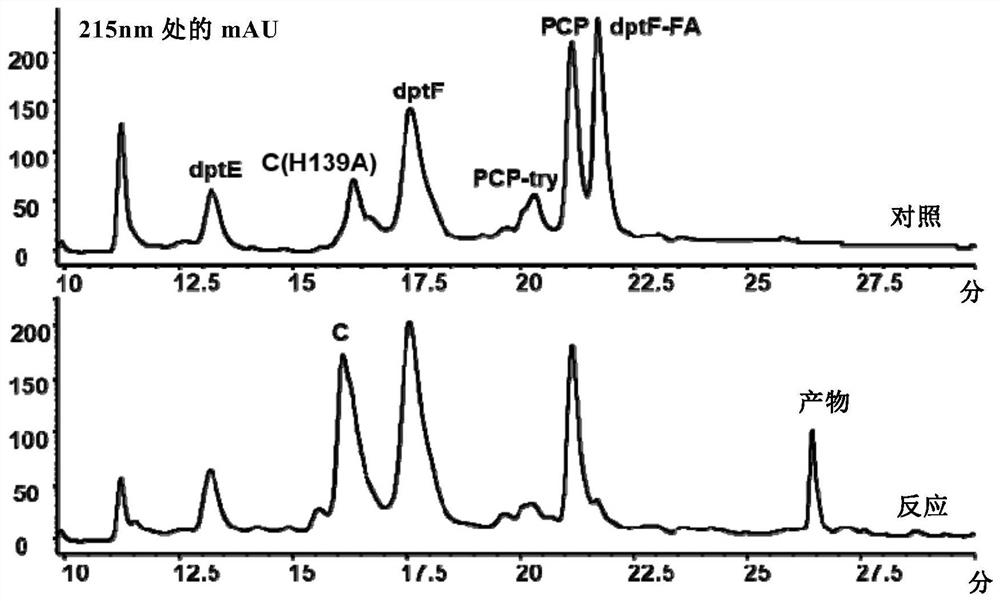
Construction of a dptc1 mutant for daptomycin biosynthesis
A technique for dptc1 and mutant proteins, applied in the field of construction of dptC1 mutants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0135] Example 1: Establishment of dptC1 in vitro enzyme activity identification and screening method
[0136] In order to construct a screening method for in vitro enzyme activity identification of dptC1, we carried out a series of work such as strain construction, multi-enzyme expression and purification, and in vitro multi-enzyme reaction system construction. The specific operation is as follows:
[0137] 1.1 Construction of Escherichia coli strains for enzyme expression and in vitro purification of enzymes
[0138] The dptC1, dptPCP1, dptE, and dptF genes of Streptomyces roseospora daptomycin fatty acid-loaded related NRPS were constructed into the pET-28a(+) vector by molecular cloning technology. The N-terminus of the protein has a his tag and is purified by Ni-NTA gravity column. The purified buffer is 50mM Tris-HCl (pH 7.6), 0.5M NaCl, 10mM MgCl 2 . The nickel column was decontaminated with washing buffer containing 20mM and 50mM imidazole successively, and final...
Embodiment 2
[0149] Example 2: Design of dptC1 small intelligent mutation library
[0150] 2.1 Homology modeling and molecular docking
[0151] Because the crystal structure of dptC1 has not yet been resolved, the crystal structure of the homologous protein CDA-C1 (41.05% sequence homology) (PDB: 4JN3) was used as a template, and the SWISSMODEL online software was used for homology modeling to obtain dptC1 simulation structure. Centering on His139, the second His catalytic site in the conserved motif "HHxxxDG" in the C domain, the model substrate channel was positioned using Schrodinger molecular simulation software. Analysis localizes residues and regions important for catalysis and substrate binding.
[0152] 2.2 Construction of dptC1 substrate pocket region mutants
[0153] According to the results of protein-substrate molecular docking, select the vicinity of the fatty acyl chain that docks into the substrate pocket The residues within are used as key residues, and mutations a...
Embodiment 3
[0157] Example 3: Cloning, expression and purification of dptC1 mutants
[0158] The dptC1 mutant was point-mutated using the wild-type pET-28a(+) vector as a template, expressed in Escherichia coli BL21(DE3), and purified in vitro using a Ni-NTA gravity column.
[0159] The whole plasmid amplification system constructed by the mutants is shown in Table 2.
[0160] Table 2 Total plasmid amplification reaction system (50 μL)
[0161]
[0162]
[0163] The PCR reaction program was: pre-denaturation at 98°C for 3 min; denaturation at 98°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 95 s, a total of 30 cycles; finally, extension at 72°C for 5 min. After amplification, the whole plasmid PCR product was detected by 1% agarose gel electrophoresis. After the PCR product was purified, the template was digested with DpnI enzyme, and then the plasmid was transformed into Escherichia coli BL21(DE3), and a single clone was picked for sequencing to verify the suc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com