High throughput detection kit of SARS-CoV-2
A sars-cov-2 and kit technology, which is applied in the detection/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of large proportion of exogenous nucleic acid data, unfavorable virus prevention and control, long time, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
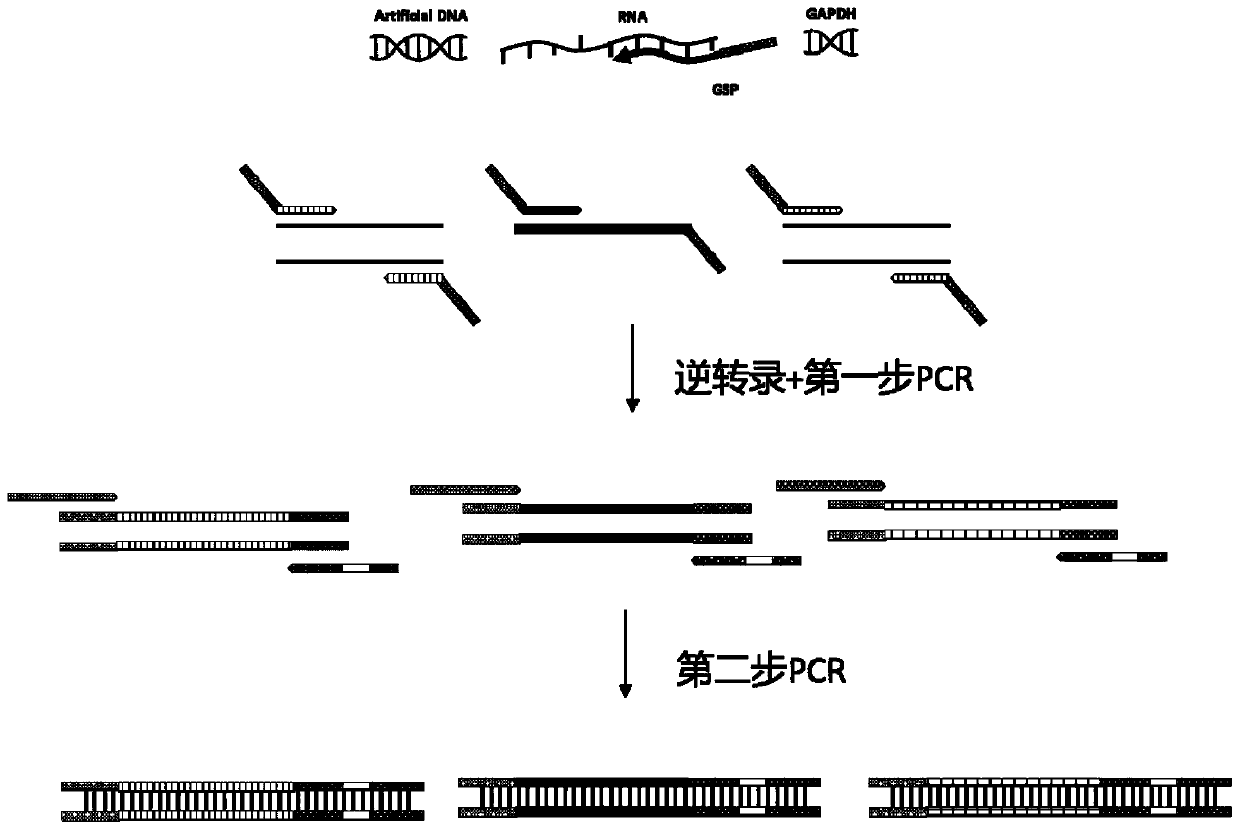
[0108] Embodiment 1, the establishment of the method for detecting SARS-CoV-2 by high-throughput sequencing
[0109] After a lot of experiments, the inventors of the present invention have established a method for detecting SARS-CoV-2 by high-throughput sequencing, which can not only sensitively detect the presence or absence of SARS-CoV-2 (qualitative), but also judge the Concentration (quantitative). Specific steps are as follows:
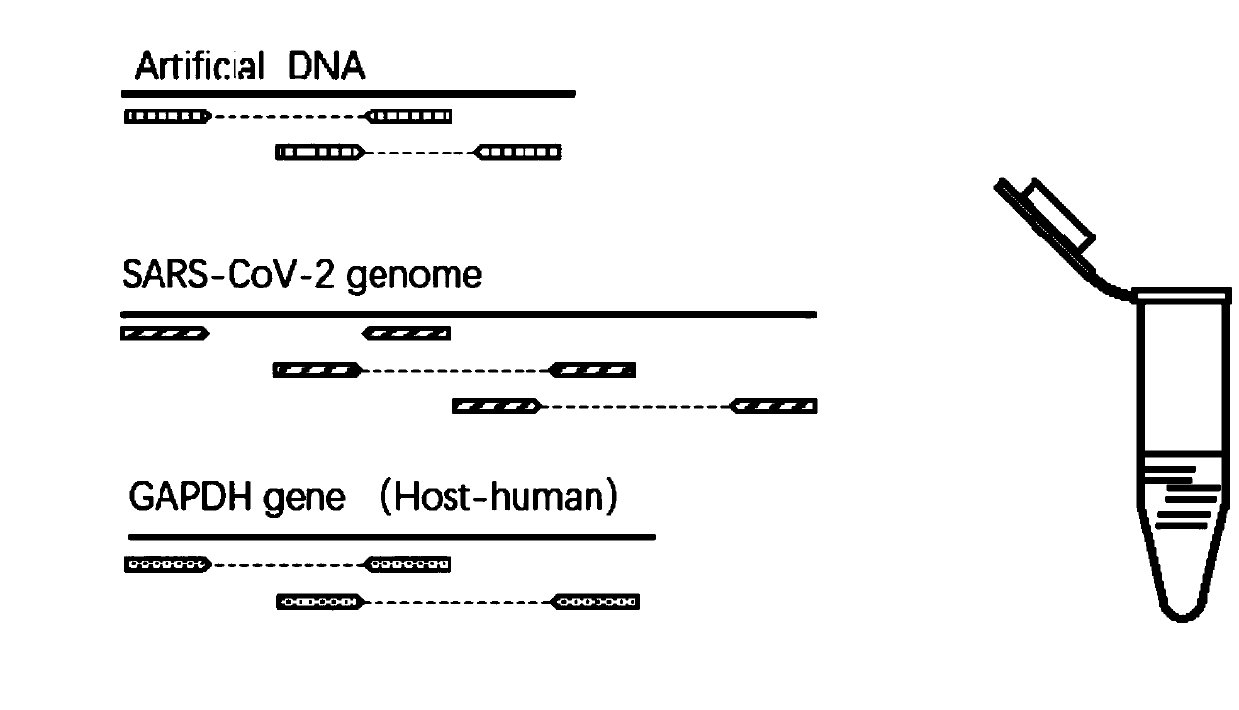
[0110] 1. Acquisition of forward primer pool and reverse primer pool
[0111] (1) Obtain the whole genome sequence of multiple SARS-CoV-2 strains, and then use the crossover design method to design primer pairs for amplifying SARS-CoV-2 (among them, design degenerate primers in the highly variable region); each The length of the target region detected by each primer pair is 300-400bp; the target region of all the primer pairs used to amplify SARS-CoV-2 can cover the entire gene of SARS-CoV-2 after splicing.
[0112] (2) According to the nucleo...
Embodiment 2
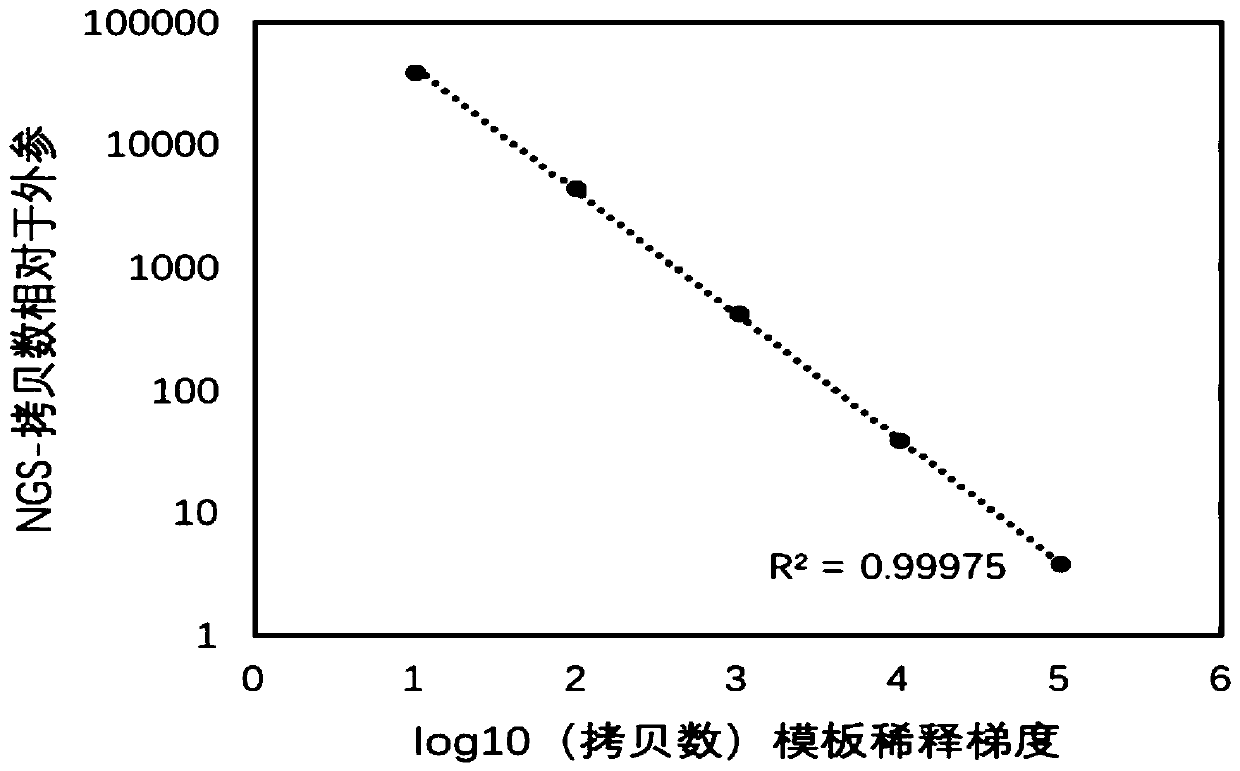
[0147] The verification of the accuracy of the method that embodiment 2, embodiment 1 establish
[0148]The method established in Example 1 was used to detect different concentrations of SARS-CoV-2, thereby evaluating the accuracy of the method established in Example 1. Specific steps are as follows:
[0149] 1. Preparation of sample 1-sample 5
[0150] 1. Take a throat swab from a patient infected with SARS-CoV-2 and extract nucleic acid.
[0151] 2. After completing step 1, take the nucleic acid and dilute it to 10 times with water to obtain a dilution of 10 -1 Nucleic acid diluent 1; then continue to dilute to obtain a dilution of 10 -2 Nucleic acid diluent 2, the dilution is 10 -3 Nucleic acid diluent 3, the dilution is 10 -4 Nucleic Acid Dilution 4 and Dilution 10 -5 Nucleic acid dilution solution 5.
[0152] 3. After step 2 is completed, qPCR is used to detect the absolute copy numbers of nucleic acid dilution 1-nucleic acid dilution 5 respectively.
[0153] The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com