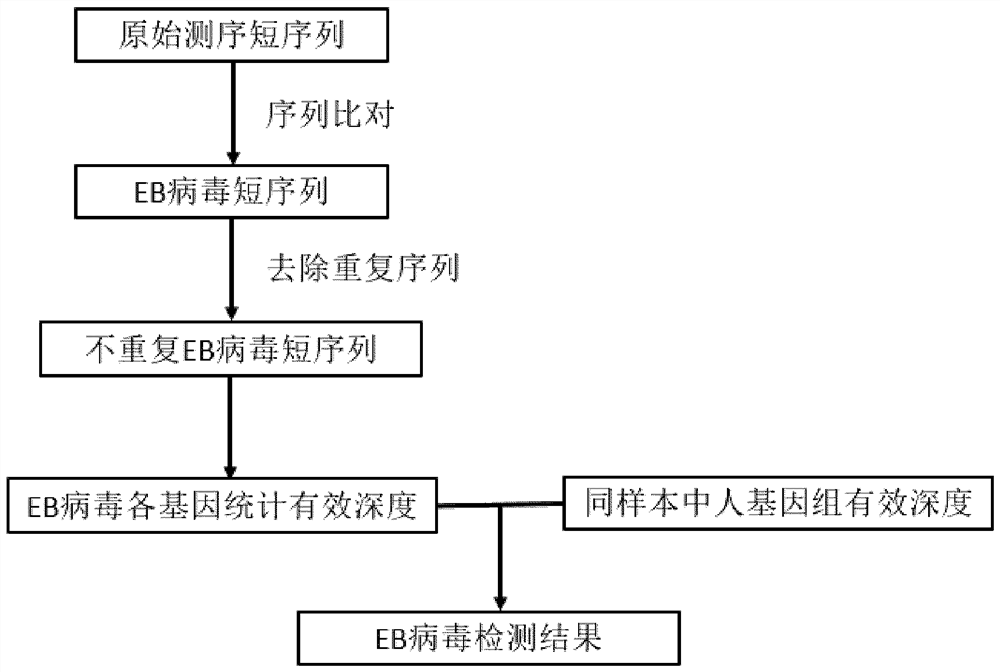
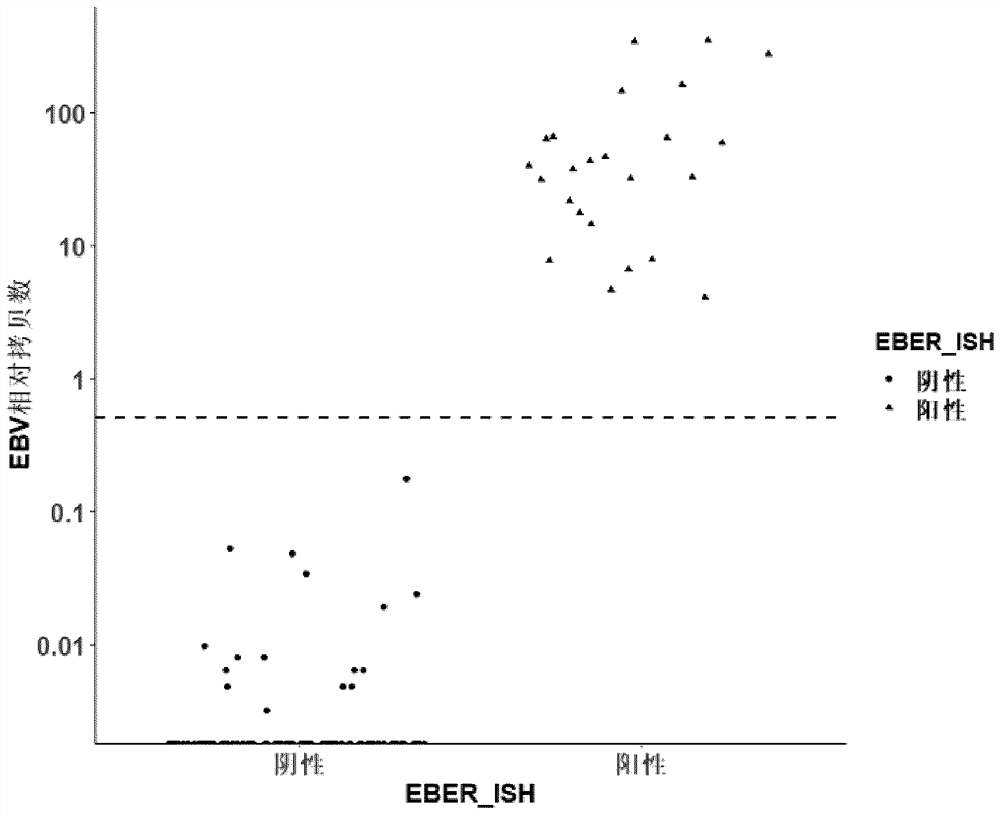
EB virus detection technology based on capture sequencing
An Epstein-Barr virus and sequencing technology, which is applied in recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve problems such as poor sample quality, large impact on test results, and inability to combine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Example Embodiment
[0052]Example 1: Proof of specificity of EB virus probe design
[0053]● Use bedtools2 (the same below v 2.25.0) software to extract the area of the probe design (that is, the area covered by the probe of the present invention) and integrate it to form a bed file;
[0054]● Use bedtools software to extract the Epstein-Barr virus reference genome sequence (GenBank ID: NC_007605.1) according to the generated bed file to form a fasta file;
[0055]● Use the internal python script to split each sequence in the fasta file to form a fastq file. The split method is: use 1bp as the step size and 100bp as the window, slide on each record, and finally generate a fastq file with a length of 100bp ;
[0056]● Use bwa v0.7.12 software to compare the split fastq file to the human reference genome (GRCH37) to generate a comparison file (bam format);
[0057]● Use the python script to view the comparison result information to verify whether the Epstein-Barr virus and the human genome have homologous regions.
[00...
Example Embodiment
[0061]Example 2: Proof of specificity of EBV-1 and EBV-2 probe design
[0062]● Use bedtools2 (the same below v 2.25.0) software to extract the corresponding ranges of the two types of EB virus in the probe design area (that is, the area covered by the probe of the present invention) and integrate them to form a bed file;
[0063]● Use bedtools software to extract the Epstein-Barr virus reference genome sequence according to their respective bed files, and form fasta files respectively;
[0064]● Use the internal python script to split each sequence in the fasta file to form a fastq file. The split method is: use 1bp as the step size and 100bp as the window, slide on each record, and finally generate a fastq file with a length of 100bp ;
[0065]● Use bwa v0.7.12 software to compare the split fastq files to the reference genome of the Epstein-Barr virus, and generate comparison files (bam format) respectively;
[0066]● Use python script to view the comparison result information.
[0067]● The final ...
Example Embodiment
[0072]Example 3: Library building, capturing and sequencing of samples
[0073]After obtaining the tissue sample, first use a mature kit on the market to extract the DNA. The DNA from tissues needs to be interrupted by ultrasonic technology, DNA digestion technology or transposase technology to form a double-stranded DNA fragment with a length of about 200 bases. Then use the corresponding kits to construct the library for DNA from different sources. After the library is built, the libraries of one to four samples are mixed, and then the Epstein-Barr virus capture probe is used to capture the Epstein-Barr virus genome fragments. The EB virus capture probe can be mixed with other probes to capture together, and the capture efficiency of the EB virus probe will not be affected. The captured library is finally sequenced by a high-throughput sequencer.
[0074]Operation steps of wet experiment:
[0075]Step 1: Build a library:
[0076]1. Reagents and consumables
[0077]a) KAPA Hyper Library Preparati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com