A method for preparing precise blg gene knockout cattle using third-generation base editors
A gene knockout and editor technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, genetic engineering, etc., can solve the problems of difficult point mutation technology, fragment deletion, limited HDR, etc., to avoid random Effects of mutations and even genomic damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1. Preparation of BLG biallelic mutant cell lines using third-generation base editors
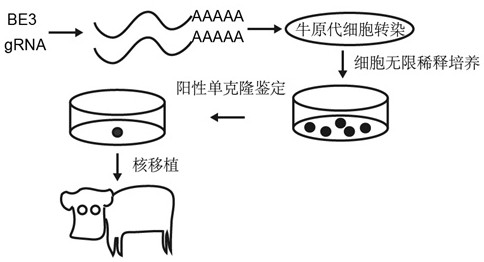
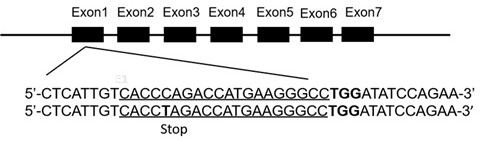
[0040] In this example, the third-generation base editor (BE3) was used to prepare BLG biallelic mutant cell lines. Considering the later industrialization and avoiding biological safety issues, the present invention does not use screening marker genes, but uses BE3 mRNA at the same time, so that BLG biallelic mutant cell lines without exogenous DNA integration can be prepared. For the technical route, see figure 1 . The target region sequence of BE3 single-base modification technology and the schematic diagram of BLG gene knockout achieved by single-base modification technology of BLG gene are shown in figure 2 .
[0041] According to the BLG gene sequence (SEQ ID No.1), the region where the base editing target sequence is located is located on exon 1 (SEQ ID No.2) of the BLG gene sequence, and the binding sequence of the target region is cacc c agaccatgaagggccTGG (posit...
Embodiment 2
[0088] Embodiment 2, the preparation of BLG biallelic knockout cattle
[0089] 1. Take mutant fibroblasts in the logarithmic growth phase and digest them with 0.25% trypsin for 5 minutes to obtain single cells.
[0090] 2. Collect the ovaries of adult Holstein cows from the slaughterhouse, wash them three times in PBS solution at 37°C, extract follicles with a diameter of 2-8mm with a needle with a diameter of 0.7mm, and recover cumulus-eggs with uniform shape and dense structure Mother cells-complexes (COCs), washed twice with maturation solution (M199+10%FBS+0.01U / mL bFSH+0.01U / mL bLH+1µg / mL estradiol), and then the cumulus-oocyte - Put 50-60 pieces / well of the complex into a four-well plate containing maturation solution, at 38.5°C, 5% CO 2 After maturing and culturing in the incubator for 18-20 hours, put the mature oocytes into the tube of 0.1% hyaluronidase and shake for 2-3 minutes, then blow gently with a glass tube to completely separate the cumulus cells from the oo...
Embodiment 3
[0099] Example 3, Analysis of BLG protein expression in the milk of BLG biallelic knockout cows
[0100] 1. SDS-PAGE detection of BLG protein in the milk of BLG biallelic knockout cows
[0101] 1. Extract the total protein from the milk produced from the first day to the seventh day of lactation of the cow to be tested (BLG bi-allelic knockout cow or wild Holstein cow).
[0102] 2. Take the total protein of the cow to be tested and the BLG protein standard (product of Sigma) obtained in step 1, respectively, and carry out SDS-PAGE.
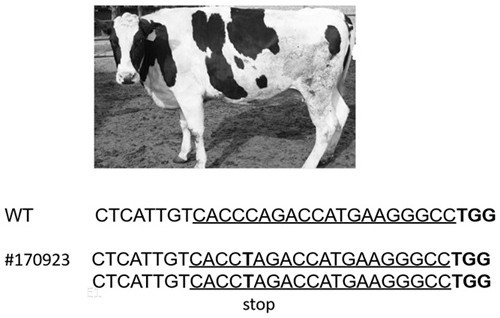
[0103] See the experimental results Figure 4 (M is protein marker, BLG is BLG protein standard). The results showed that the expression of BLG protein was not detected in the total protein of BLG biallelic knockout cows. It can be seen that there is no BLG protein in the milk produced by BLG biallelic knockout cows. Not only the BLG gene was knocked out at the DNA level, but also Levels are also effective in removing BLG.
[0104] 2. Western ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com