Cancer subtype classification method based on multi-omics integration
A classification method and omics technology, applied in the field of cancer subtype classification based on multi-omics integration, can solve the problems of large error, no consideration of sample similarity bias, omics data weight, poor accuracy of cancer subtype classification results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
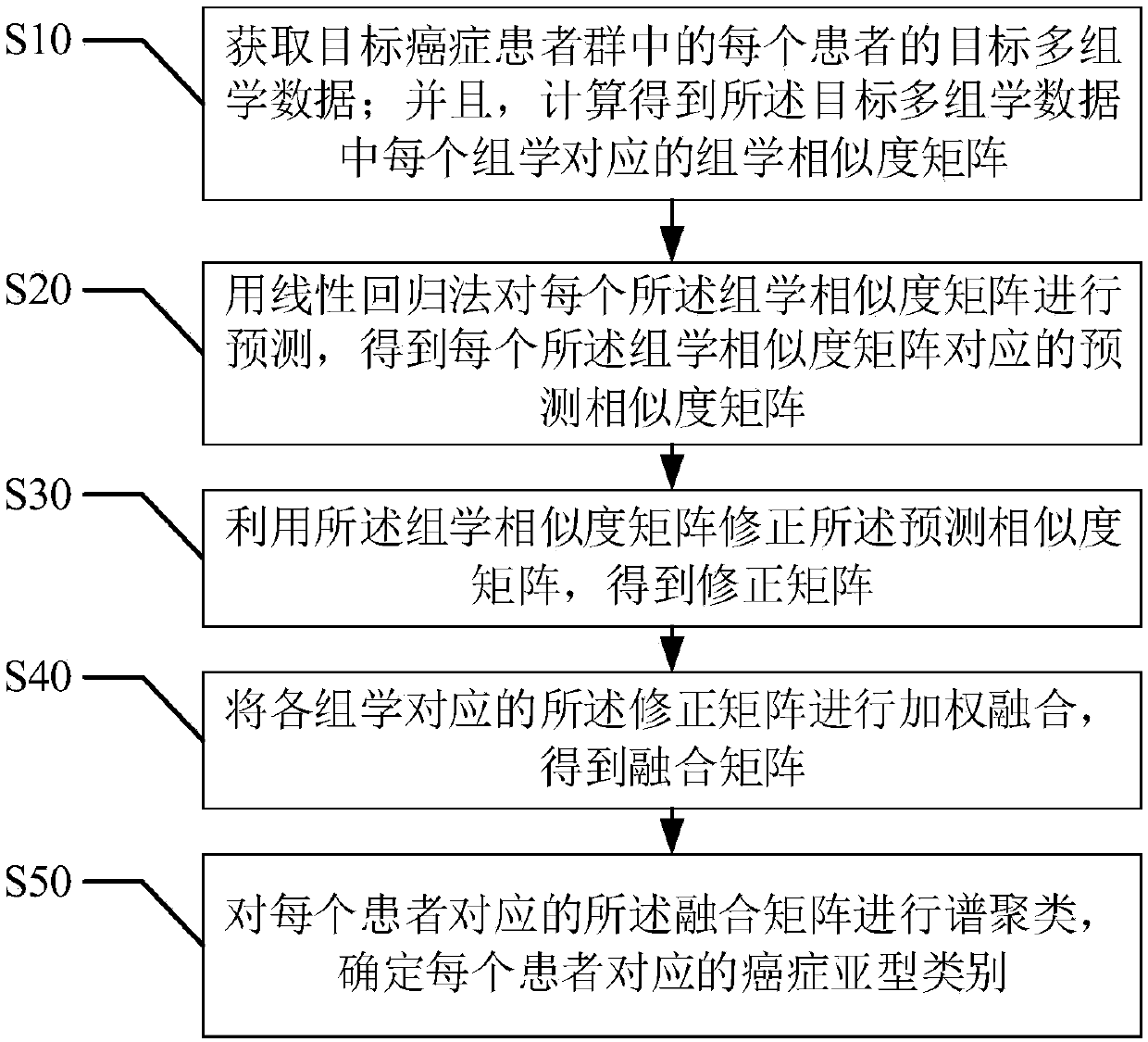
[0066] refer to figure 2 , the first embodiment of the present invention provides a cancer subtype classification method based on multi-omics integration, including:
[0067] Step S10, acquiring target multi-omics data of each patient in the target cancer patient group; and calculating an omics similarity matrix corresponding to each omics in the target multi-omics data;
[0068] As mentioned above, the omics similarity matrix is M k ; among them, M k Can be expressed in the following form:
[0069]
[0070] As mentioned above, the target cancer patient group is a collection of batch cancer subtype classification for all patients in the group that need to be analyzed. The target cancer patient group includes pathological data (physical and chemical index data, biochemical test results, etc.) of patients with the same type of cancer but the same and or different conditions.
[0071] As mentioned above, the target cancer patient group includes multiple patients with th...
Embodiment 2
[0093] refer to image 3 , the second embodiment of the present invention provides a cancer subtype classification method based on multi-omics integration, based on the above figure 2 In the first embodiment shown, the step S20 of "predicting each of the omics similarity matrices using a linear regression method to obtain a predicted similarity matrix corresponding to each of the omics similarity matrices" includes:
[0094] Step S21, based on the linear regression method, respectively use the omics similarity matrix corresponding to each omics of the target multi-omics data of each patient as the target matrix, and use the omics similarity matrix corresponding to other omics to pair performing linear regression prediction on the target matrix, respectively obtaining predicted values corresponding to data in each of the target matrices of the target multi-omics data, and obtaining each of the omics similarity matrices containing the predicted values The corresponding predi...
Embodiment 3
[0111] refer to Figure 4-5 , the third embodiment of the present invention provides a cancer subtype classification method based on multi-omics integration, based on the above figure 2 In the first embodiment shown, the step S10, "acquire the target multi-omics data of each patient in the target cancer patient group; and calculate the group corresponding to each omics in the target multi-omics data Learning similarity matrix" includes:
[0112] Step S11, determining the target multi-omics data of each patient in the target cancer patient group, and performing mean interpolation on data corresponding to missing omics;
[0113] As mentioned above, in the target multi-omics data, there are multiple omics, but due to the large number of patients, not every patient has completed the test of each omics, and there may be missing tests, resulting in some patients missing In the case that a certain omics cannot be calculated, it is necessary to interpolate the missing omics of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com