Applications of circRRM2B gene as new target in screening Vemurafenib-resistant melanoma treatment drugs
A fenib-resistant, melanoma technology, applied in the field of bioengineering, can solve the problem of no biological function of circRRM2B, and achieve the effect of large toxic side effects and high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
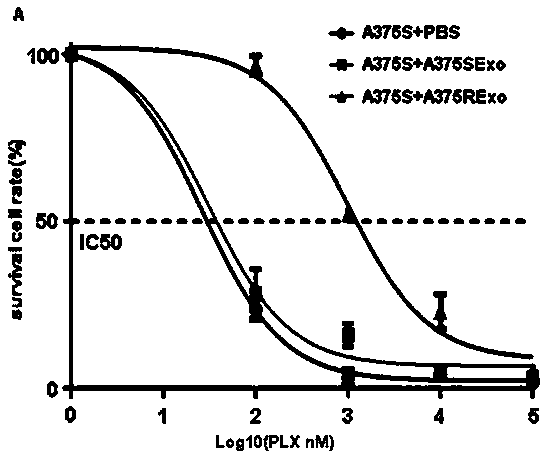
[0017] Example 1 Co-cultivation of exosomes derived from drug-resistant strains and sensitive strains can increase the resistance of sensitive strains to vemurafenib.
[0018] 1-1) Multiple strains of melanoma cells were cultured, IC50 values were measured after vemurafenib treatment, and sensitive strains (A375S) were selected. The corresponding drug-resistant strain (A375R) was established by using the drug concentration increasing method.
[0019]1-2) Cultivate drug-resistant and sensitive strains, collect the cell supernatant, centrifuge at 110,000×g for 2 hours, remove the supernatant, and resuspend the pellet in PBS. Centrifuge at 110,000×g for 70 minutes at 4°C. The pellet was resuspended in PBS, centrifuged at 110,000×g for 70 minutes at 4°C (twice), and the obtained pellet was resuspended in PBS and stored at -80°C. The morphology of exosomes was observed by transmission electron microscopy. Exosome marker proteins CD63 and CD81 were detected by WB.
[0020] 1-3...
Embodiment 2
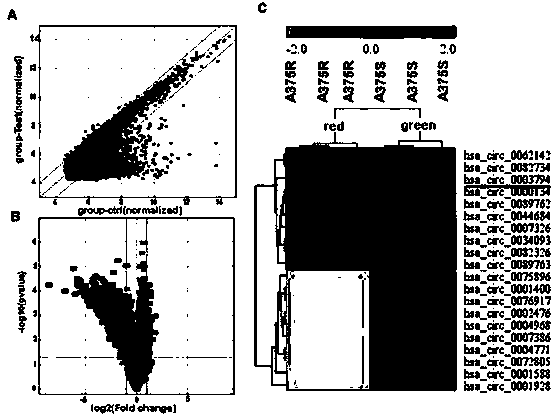
[0022] Example 2 The circRNA chip screened out the 10 most differentially expressed circRNAs in the exosomes of sensitive strains and drug-resistant strains.
[0023] 2-1) Digest the total RNA in exosomes with RNase R enzyme to remove linear RNA and enrich circular RNA. Then, the labeled cRNA was hybridized to Arraystar Human circRNA Array V2 (8x15K, Arraystar) by random primer method, the chip was washed after incubation at 65°C for 17 hours, and the array was scanned with Agilent G2505C scanner.
[0024] 2-2) The collected array images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed using the R software limma package. Statistically significant differentially expressed circRNAs were identified between the two groups by volcano plot screening. Differentially expressed circRNAs between two samples were identified by the Fold Change filtering method. A hierarchical clustering appr...
Embodiment 3
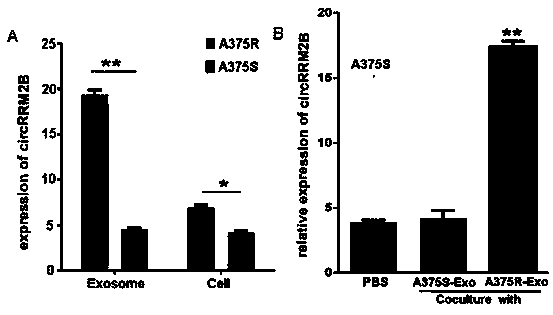
[0027] Example 3 qRT-PCR verification found that the expression of circRRM2B was significantly increased in drug-resistant strains and their exosomes. The expression of circRRM2B in sensitive strains was significantly increased after the exosomes secreted by drug-resistant strains were co-cultured with sensitive strains.
[0028] 3-1) Extract RNA with TRIZOL, measure RNA purity and concentration, reverse transcribe RNA into cDNA, and configure Realtime PCR reaction systems for all cDNA samples. qRT-PCR was used to detect the expression of circRRM2B in drug-resistant strains, drug-resistant strain exosomes, sensitive strains, and sensitive strain exosomes.
[0029] 3-2) Exosomes derived from sensitive strains and drug-resistant strains were co-cultured with sensitive strains for 48 hours, respectively, and the expression of circRRM2B in cells was detected by qRT-PCR.
[0030] Depend on image 3 It can be seen that the expression of circRRM2B was significantly increased in dru...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com