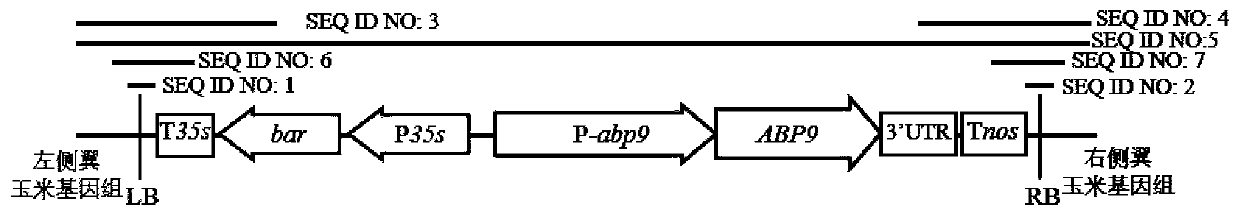
Nucleotide sequence for detecting maize plant NAZ-4 and detection method of nucleotide sequence
A sequence, technology of corn, applied in the field of plant biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
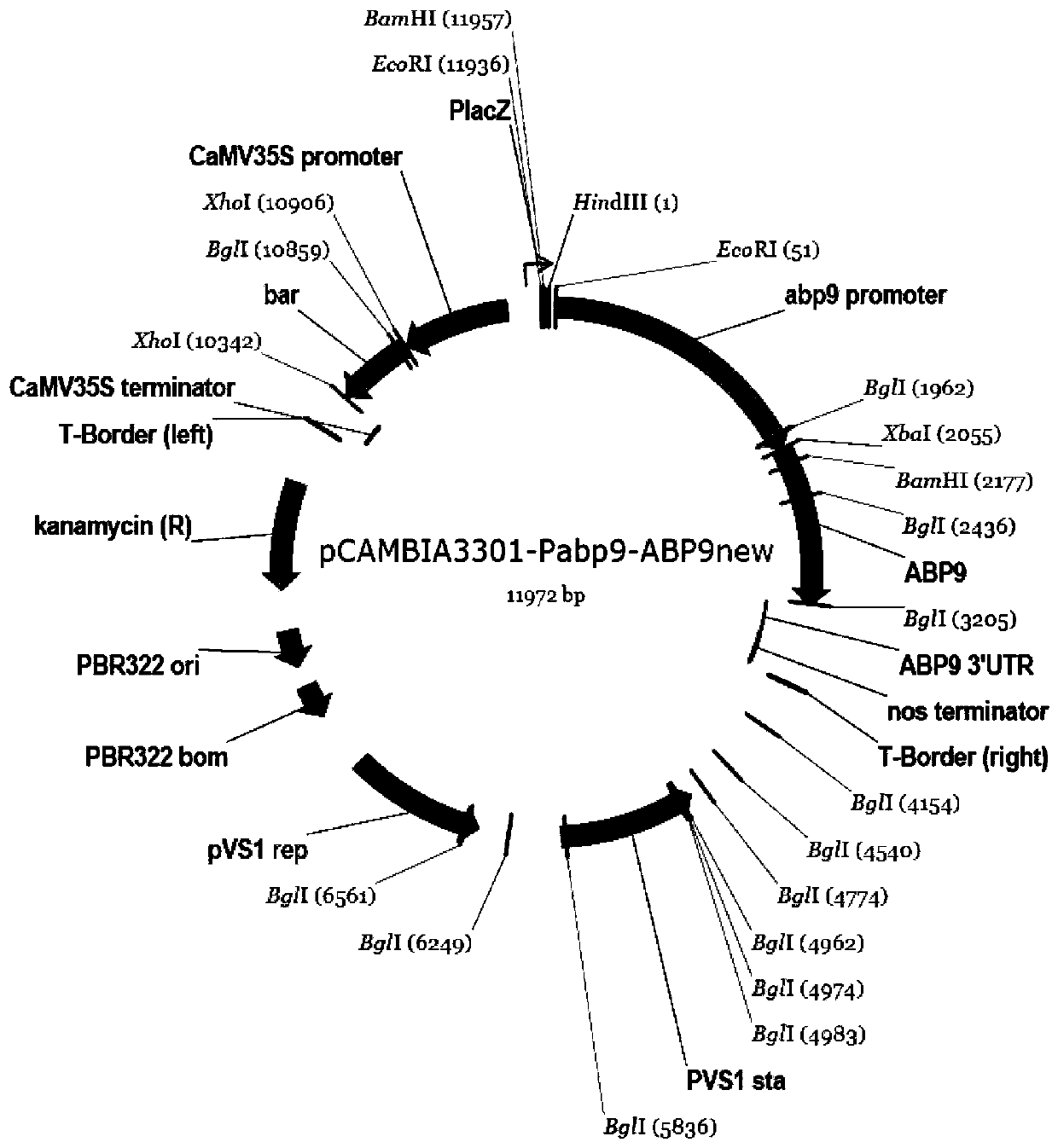
[0111] The construction of embodiment 1 transformation vector
[0112] A forward primer containing a HindIII anchor site was designed at the 5' end of the ABP9 promoter Pabp9 sequence, and a reverse primer containing a HindIII and SpeI anchor site (from outside to inside) was designed at the 3' end, and the maize genomic DNA was used as a template for PCR amplification. Pabp9 was increased; the PCR product was cloned into the pCAMBIA3301 vector by HindIII digestion to obtain the pCAMBIA3301-Pabp9 intermediate vector. The ABP9 cDNA cloning vector T-easy-ABP9 was digested with XhoI to make up, and then digested with XbaI. The recovered small fragment was ligated with the pBI121 vector digested by XbaI and Ecl136II to obtain the intermediate vector pBI121-35S-ABP9. pCAMBIA3301-Pabp9 was digested with SpeI and then digested with AflII to recover a large fragment; pBI121-35S-ABP9 was digested with XbaI and then digested with AflII to recover a small fragment; the two recovered frag...
Embodiment 2
[0114] Embodiment 2 Maize genetic transformation
[0115] The method used for transforming corn is the Agrobacterium-mediated method, and its operation procedure is as follows:
[0116] (1) select the corn ears of pollination 10-13 days, and peel off the immature embryos (1.5-2.0mm) of moderate size therefrom;
[0117] (2) Use an inoculation loop to pick a ring full of Agrobacterium LBA4404 (containing the expression vector) from the YEP plate grown at 19°C for three days, suspend it in the infection culture medium, room temperature, 75rpm, 2-4h, to OD 550 = 0.3-0.5, infecting detached immature embryos;
[0118] (3) The immature embryos that have been infected are transferred to the co-culture medium, and cultured in the dark at 20°C for 3 days;
[0119] (4) Three days later, the immature embryos were transferred to the recovery medium, and cultured in the dark at 28°C for 7 days;
[0120] (5) transfer the immature embryos to the selection medium after the recovery culture,...
Embodiment 3
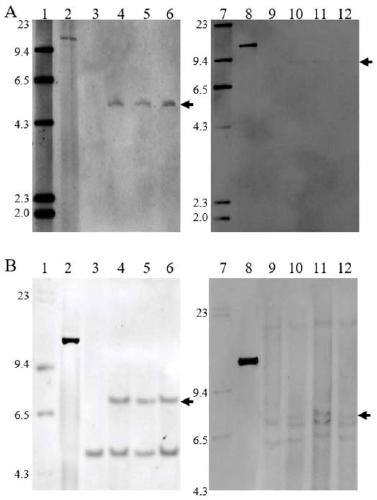
[0123] The screening of embodiment 3 transformants
[0124] (1) Molecular detection of bar and ABP9 target gene was carried out on the transformed seedlings obtained by transformation. According to gene sequence design PCR primer pair, primer sequence is respectively SEQ ID NO:16 and SEQ ID NO:17 and SEQ ID NO:18 and SEQ IDNO:19. Genomic DNA was extracted from transformed seedling leaves and amplified according to the following PCR parameters:
[0125] reaction system:
[0126]
[0127] Reaction procedure:
[0128]
[0129]
[0130] Positive transformed seedlings were screened according to the expected amplified fragment size of 672 bp of the bar gene and 582 bp of the expected amplified fragment size of the ABP9 gene.
[0131] (2) Cross the positive plants with Zheng 58 and backcross once to harvest BC 1 f 1 seed. to BC 1 f 1 The plants were identified for glufosinate-ammonium herbicide, and the transformants NAZ-1~NAZ-6 with excellent resistance were selecte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com