Genetic engineering bacterium for synthesizing pyruvic acid and D-alanine as well as construction method and application thereof
A technology of genetically engineered bacteria and pyruvate, applied in the field of genetic engineering, can solve the problems of consumption product pyruvic acid and low yield of pyruvate, and achieve the effects of simple construction method, good application prospect and easy use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Construction of gene knockout homology arm fragments
[0035] Table 1
[0036]
[0037]
[0038] According to the sequence information of E.coli BL21(DE3), design the primers shown in the table above, and use the above primers to use the E.coli BL21(DE3) genome as a template to amplify btsT, cstA, aceEF, pflB, poxB, pps, The upstream and downstream homology arm fragments of each gene of ldhA were respectively fused by Overlapping PCR to obtain fragments btsT12, cstA12, aceEF12, pflB12, poxB12, pps12, and ldhA12.
Embodiment 2
[0040] Construction of pTarget plasmid
[0041]Select the N20 region (20bp base sequence used to target the gene to be knocked out in the CRISPR / Cas9 system) on each gene to be knocked out, design primers, use pTarget as a template, and replace the N20 region of the template by PCR. The PCR product was transferred into E.coli JM109, and the plasmids were extracted to obtain 7 plasmids, pTarget-btsT, pTarget-cstA, pTarget-aceEF, pTarget-pflB, pTarget-poxB, pTarget-pps, pTarget-ldhA.
Embodiment 3
[0043] gene knockout
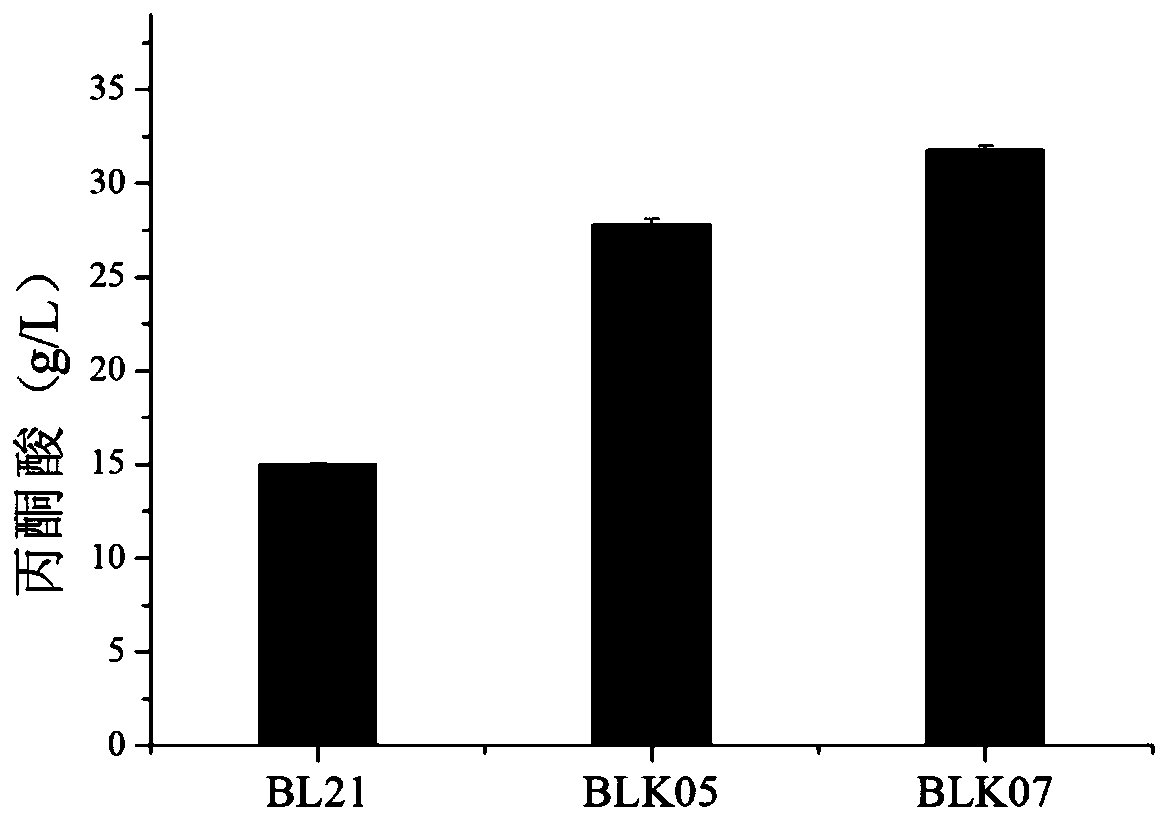
[0044] The pCas9 plasmid was transformed into E.coli BL21 (DE3) to obtain the host BL21 (pCas9) expressing Cas9 protein, and then the pTarget-aceEF plasmid and the fragment aceEF12 were electrotransformed into BL21 (pCas9) to obtain BLK01 (pCas9) with the aceEF gene knocked out , eliminate the pTarget-aceEF plasmid, electrotransfer the pTarget-pflB plasmid and the fragment pflB12 into BLK01 (Cas9), knock out the pflB gene, and knock out each gene to be knocked out in sequence according to this method, and finally eliminate the pTarget and pCas9 plasmids to obtain aceEF, pflB, poxB, pps, ldhA gene knockout BLK05 and btsT, cstA, aceEF, pflB, poxB, pps, ldhA gene knockout BLK07. BLK07 gene knockout colony PCR verification results are as follows figure 1 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com