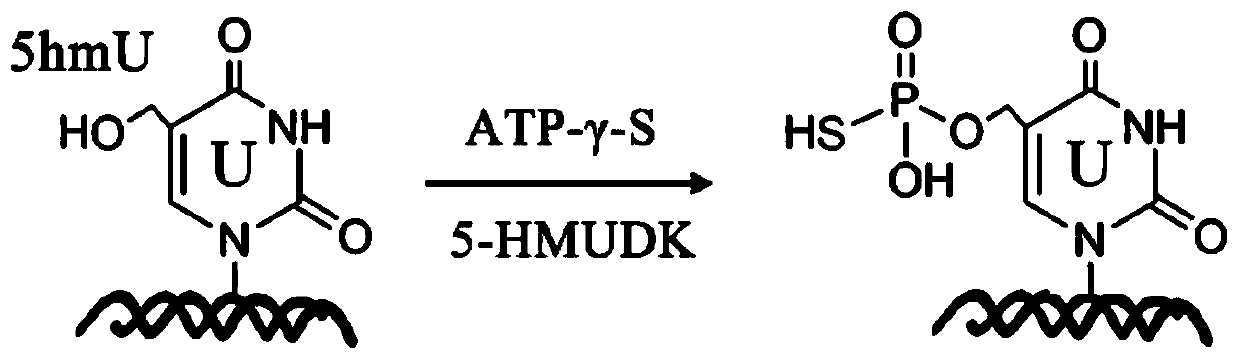
Specific marking method for 5-hydroxymethyluracil on DNA (deoxyribonucleic acid)
A technology of hydroxymethyluracil and phosphorothioatemethyluracil, which is applied in the field of specific labeling of 5-hydroxymethyluracil on DNA, can solve the problem of large demand for detection samples, harsh chemical reaction conditions, and low reaction efficiency. Low-level problems, to achieve the effect of improving accuracy and sensitivity, easy to obtain reaction reagents, and simple reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Using the specific labeling method for 5-hmU of the present invention to realize the distinction of different bases.
[0044] Prepare dsDNA oligonucleotide sequences containing different bases (including restriction endonuclease NcoI site 5'-CCAXGG-3', X=T, U, hmU and fU), and catalyze the primer synthesis by DNA polymerase in vitro For extension, dsDNA model substrates containing sites of different base pairs (5hmU:A, 5hmC:G, 5fC:G, 5fU:A, U:A or T:A) were prepared.
[0045] Wherein, in order to form 5hmU:G-containing dsDNA, the 5-hmU:A-containing dsDNA product was incubated with a G template to perform a strand displacement reaction. Product characterization in model DNA labeling reactions by 5-HMUDK and SH reaction reagents. Such as figure 2 Shown is the melting curve of the DNA sample cleaved by the restriction endonuclease NcoI, and it can be seen that the reaction product 5psmU can block the cleavage of DNA by NcoI.
[0046] 5-HMUDK thiophosphorylat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com