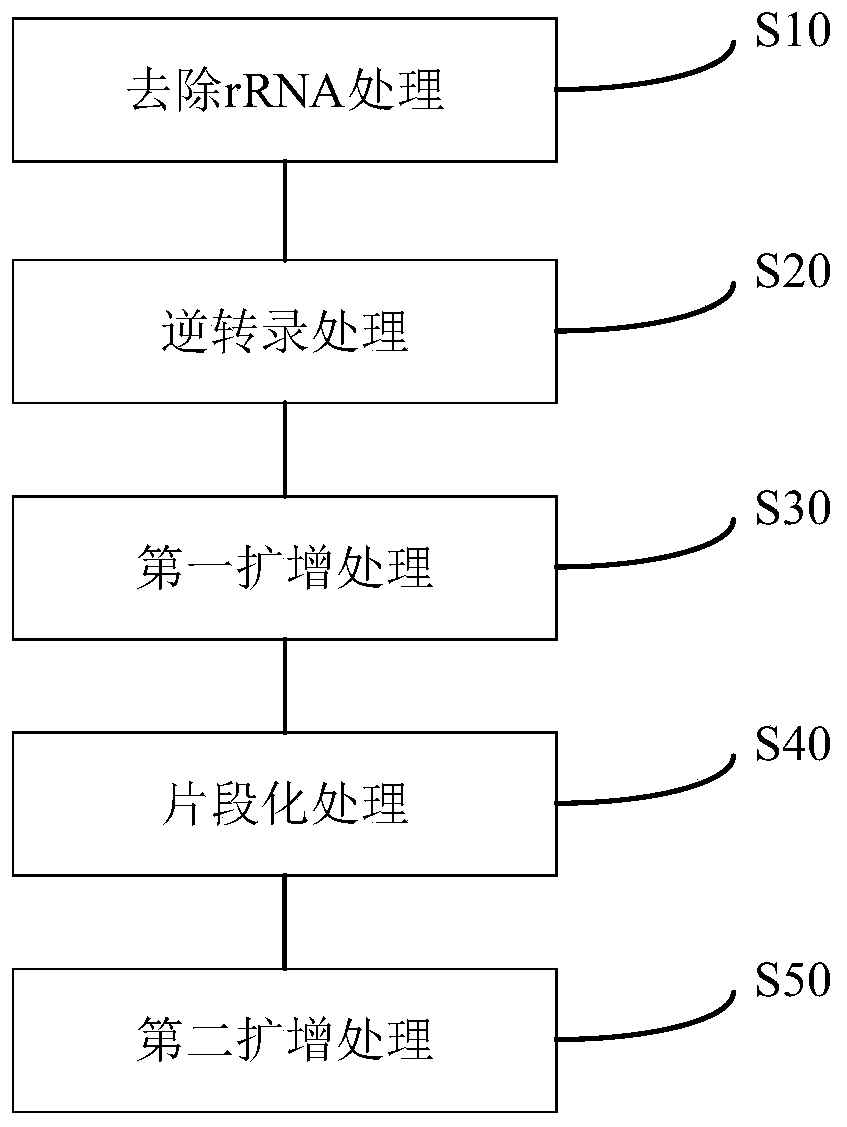
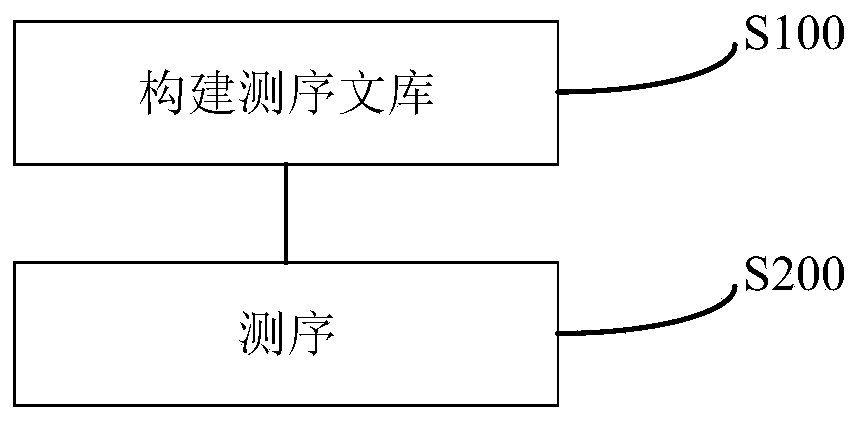
Method for constructing a DNA library and application thereof
A technology for DNA library and end repair, which is applied in the kit for constructing DNA library and in the field of constructing DNA library, which can solve the problems of long time for library construction, low conversion efficiency, and method needs to be improved, and achieve high efficiency of end repair and tailing , low preference, shortened reaction time and the effect of reaction flow
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] In this embodiment, the reaction conditions for end repair and end protrusion adenylation are optimized, as follows:
[0065] Since adenylation enzymes usually show a certain preference, DNA fragments with pyrimidine (dT or dC) at the 3'end are more likely to be adenylated than DNA fragments with purines (dA or dG) at the 3'end. Incomplete adenylation will lead to The blunt-end ligation reaction between DNA fragments affects the genome sequence assembly of the sequencing data. In order to improve this, on the basis of the polymerase basic buffer system, the pH was increased to 8.8, and the magnesium ion concentration was adjusted to 10mM to form a modified buffer suitable for one-step end repair and A-tailing (10mM Tris-HCl, 10mM MgCl2, 50mM KCl, 0.1% Triton-x-100, PH8.8), as shown in Table 1 and figure 1 As shown, compared with the existing blue buffer (Blue buffer), the reaction efficiency of terminal adenylation is improved and preference is reduced.
[0066] In this exa...
Embodiment 2
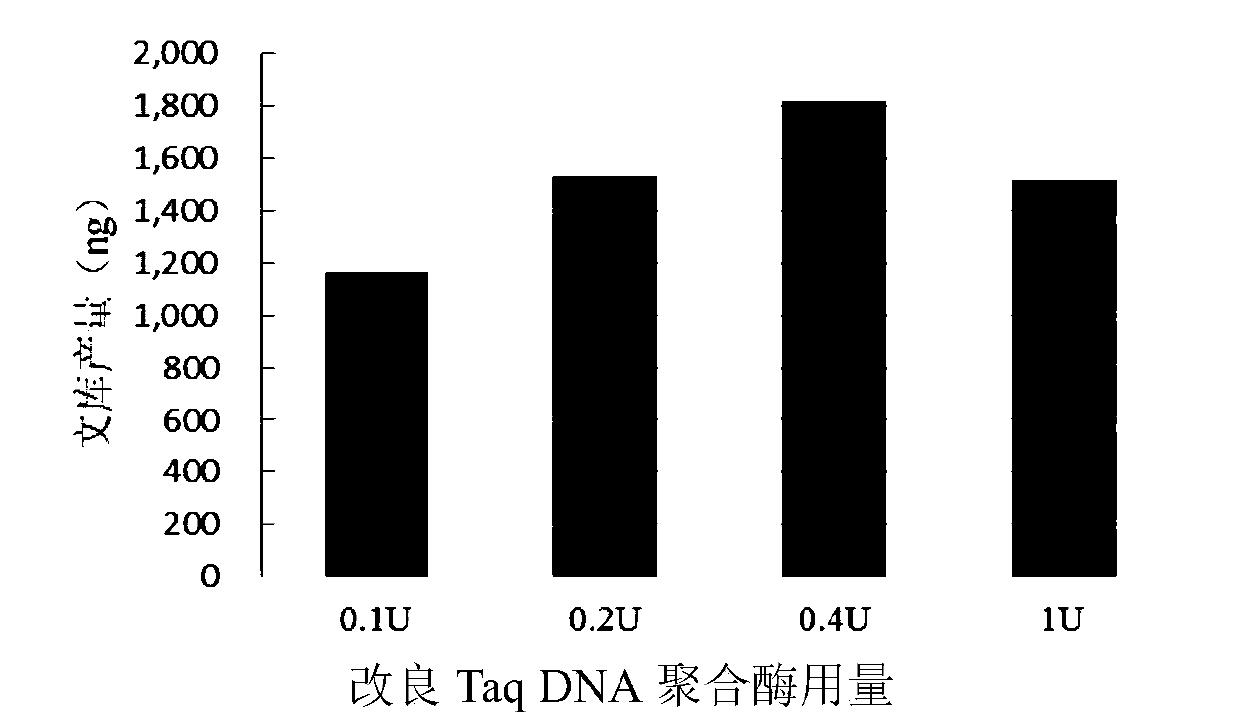
[0072] Using the modified buffer of Example 1, the influence of the temperature and time conditions of the adenylate tailing reaction on the yield of the library was explored, the amount of enzyme, and the reaction time and temperature were as follows: image 3 with 4 As shown, specifically, the concentrations of the modified Taq DNA polymerase are 0.1U, 0.2U, 0.4U, and 1U, respectively, and the reaction time and temperature of the adenylate tail reaction are 68 degrees Celsius, 10 minutes; 68 degrees Celsius, 30 minutes ; 72 degrees Celsius, 10 minutes; 72 degrees Celsius, 30 minutes; 75 degrees Celsius, 10 minutes; 75 degrees Celsius, 30 minutes. At the same time, 100ng digested DNA fragments were used as substrates to explore the usage of modified Taq DNA polymerase.
[0073] The result is image 3 with 4 Shown in image 3 In, the modified buffer and modified Taq DNA polymerase of Example 1 were used in one-step end repair and adenylate addition reaction. The amount of substrat...
Embodiment 3
[0075] In the modified buffer of Example 1, add interruption buffer (0.1×DnaseI Buffer, 1×BlueBuffer, 50mM Nacl, 0.1% TritonX-100, 120ng sso7d), and use NA12878 standard substrate to build in this buffer system. The library is evaluated for the compatibility of DNA fragmentation enzymes. The amount of substrate and library yield are shown in Table 2.
[0076] Table 2
[0077]
[0078] The conditions for interrupting the integration of library construction are 37°C for 10 minutes and 72°C for 20 minutes. Reactions 2 and 3 are repetitions of reaction 1, and the results are as follows Figure 5 Shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com