Ranking system for immunogenic cancer-specific epitopes
An antigen-determining and immunological technology, applied in the direction of cancer antigen components, specific peptides, immunoglobulins, etc., can solve problems such as impact prediction and consideration of specific cancer antigens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
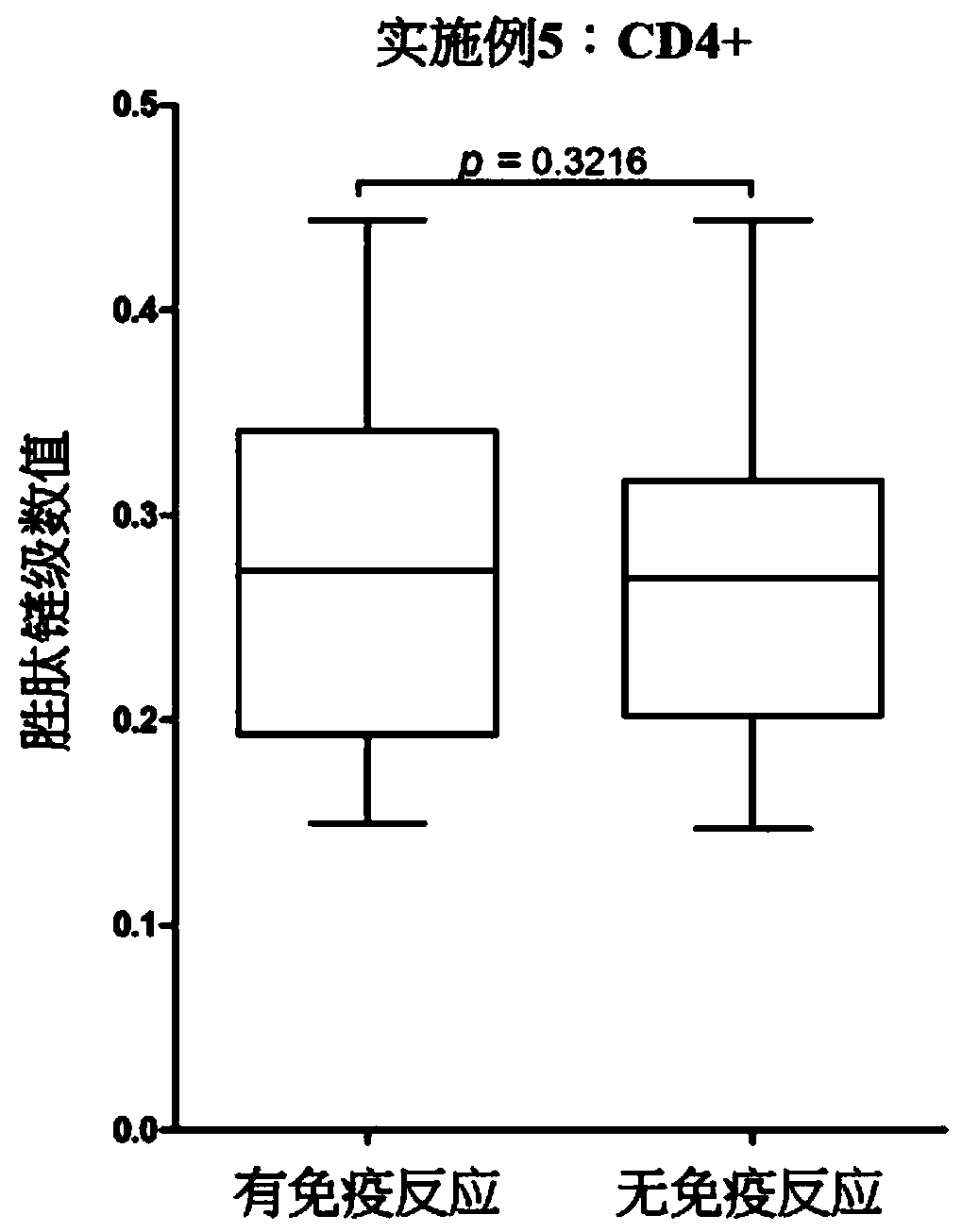
[0061] Example 1: Using the binding affinity (binding affinity) and binding stability (binding stability) between the peptide chain and the major histocompatibility complex type I (MHC class I) to predict the type of the major histocompatibility complex for the peptide chain A situation presented.
[0062] For a peptide chain to be an antibody, the peptide chain needs to be presented on the cell surface by the major histocompatibility complex and then recognized by immune cells. The above process includes: the peptide chain is presented to CD4+ T cells by the major histocompatibility complex type II (MHC class II) in the antigen-presenting cell, and the peptide chain is presented to the CD4+ T cell by the major histocompatibility complex type I in the antigen-presenting cell. Presentation to CD8+ T cells and peptide chains are presented to CD8+ T cells by major histocompatibility complex type-1 in tumor cells. In this example, we construct a model with selected features to pr...
Embodiment 2
[0064] Example 2: Using the binding affinity between the peptide chain and the major histocompatibility complex type I and the expression level of the gene to which the peptide chain belongs to predict the situation that the peptide chain is presented by the major histocompatibility complex type I.
[0065] In addition to the peptide's ability to bind to major histocompatibility complex type 1, the degree to which the peptide is represented is also important for whether the peptide is represented. In this example, we constructed a model based on the expression of the gene to which the peptide chain belongs and the binding affinity between the peptide chain and the major histocompatibility complex type 1 to predict that the peptide chain is bound by the major histocompatibility complex type A situation presented.
[0066] We calculated the binding affinity between the peptide chain and major histocompatibility complex type 1 in the manner disclosed in Example 1, and calculated ...
Embodiment 3
[0067] Example 3: Using the binding affinity between the peptide chain and the major histocompatibility complex type 1 and the expression of the protein to which the peptide chain belongs to predict the situation that the peptide chain is presented by the major histocompatibility complex type 1.
[0068] In addition to the binding capacity of the peptide chain to MHC type 1, the amount of the peptide chain represented also affects the amount of the peptide chain represented by the MHC. In this example, we used the binding affinity between the peptide chain and major histocompatibility complex type 1 and the performance of the peptide chain as the selected features to construct a method for predicting that the peptide chain is major histocompatibility A model of sexual complex type-presentation situations.
[0069]We constructed a model to predict peptides based on two properties (peptide binding affinity and peptide expression, respectively) that affect the binding capacity an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com