Method and device for predicting drugability of target ppis based on protein interaction network
A protein interaction and prediction method technology, applied in the field of bioinformatics, to achieve the effect of eliminating false positive interactions, calculating results in line with objective reality, and reducing false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
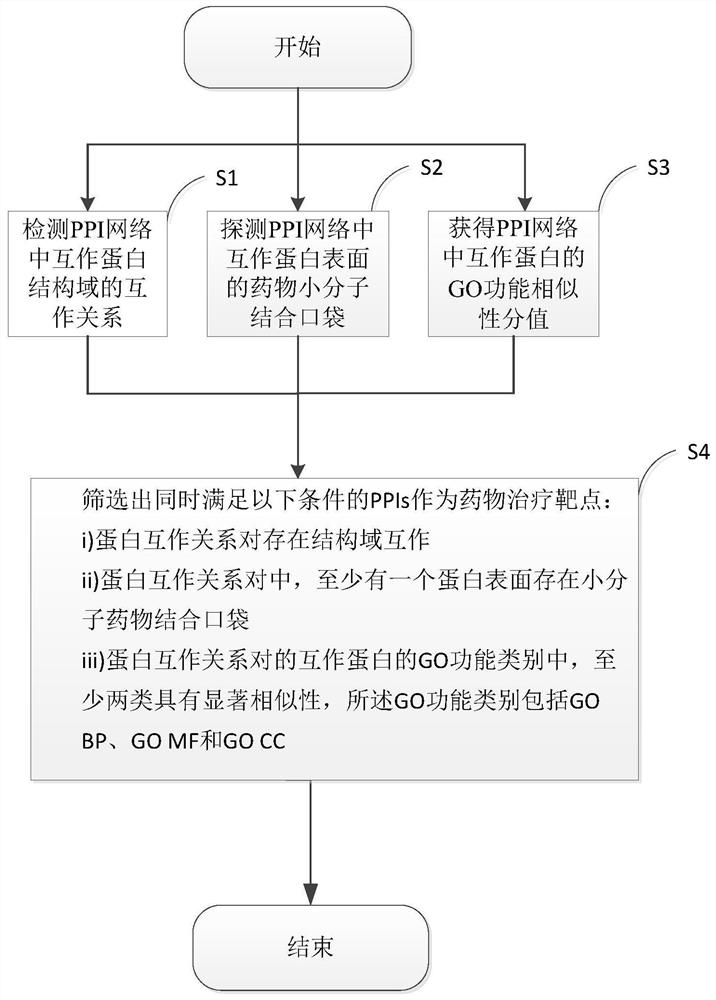
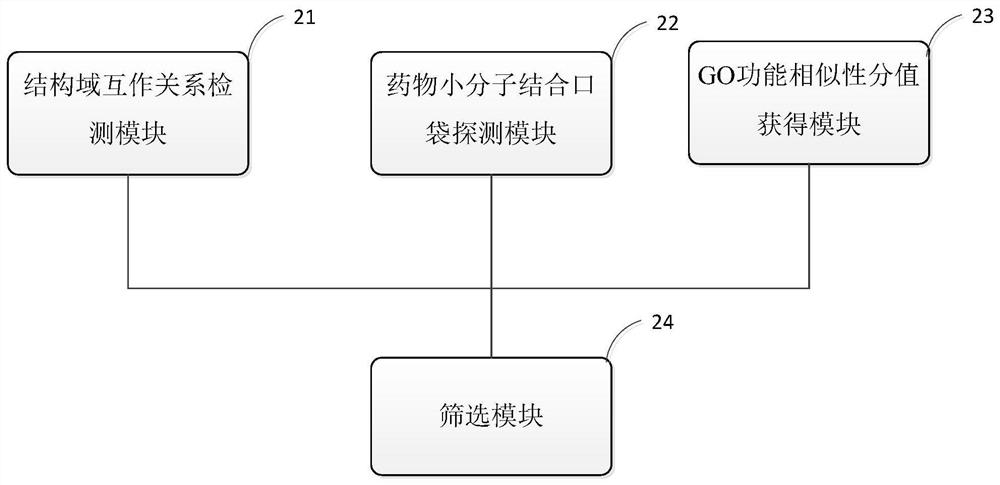
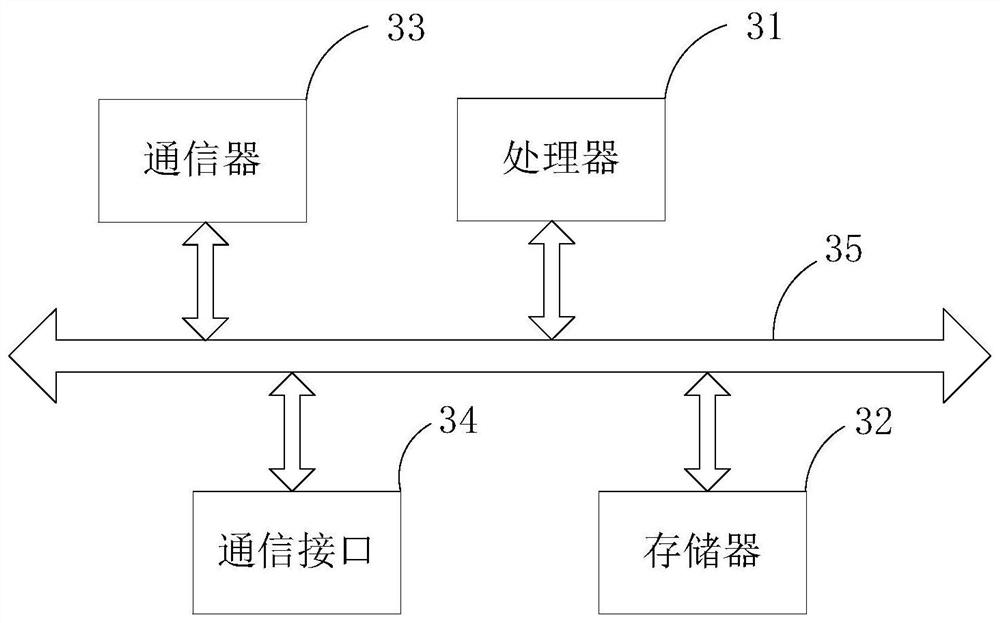
[0038] Embodiments of the present invention are described below through specific examples, and those skilled in the art can easily understand other advantages and effects of the present invention from the content disclosed in this specification. The present invention can also be implemented or applied through other different specific implementation modes, and various modifications or changes can be made to the details in this specification based on different viewpoints and applications without departing from the spirit of the present invention.
[0039] In addition, it should be understood that one or more method steps mentioned in the present invention do not exclude that there may be other method steps before and after the combined steps or other method steps may be inserted between these explicitly mentioned steps, unless otherwise There are instructions; it should also be understood that the combined connection relationship between one or more steps mentioned in the present...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com