Modified t cells and methods of their use
A technology of lymphocytes and cell receptors, applied in the field of modified T cells and their use, can solve the problem of low cell persistence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0239] Example 1: Knockout of CD3ζ in Jurkat T cells and primary T cells, and transduction of CD3ζ knockout cells with CAR
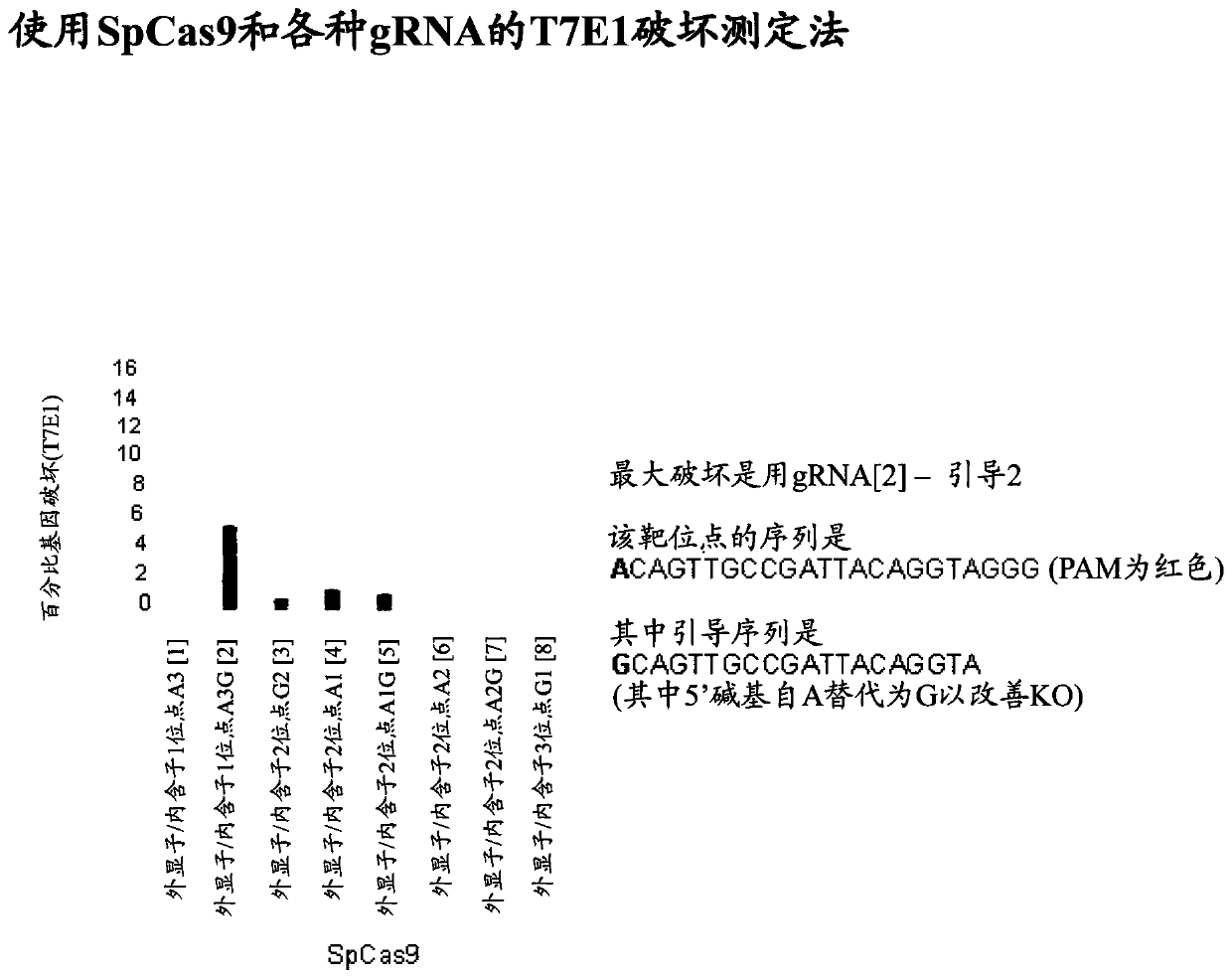
[0240] To achieve gene knockout using CRISPR, guidelines were developed targeting various coding sequences of CD3ζ, targeting CRISPR-mediated gene disruption by generating insertions and / or deletions in the targeted genomic sequence, causing loss of expression. frameshift mutation.
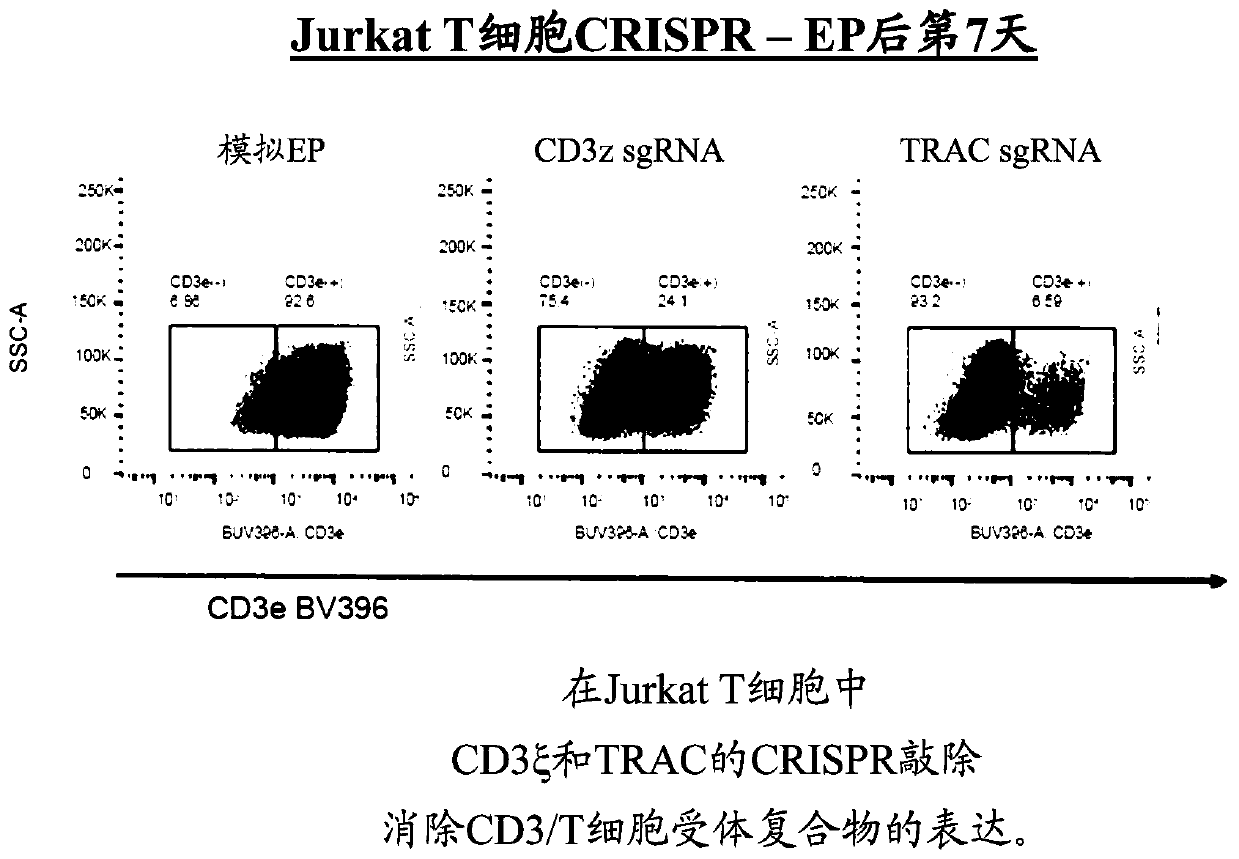
[0241] Knockdown of CD3ζ or T cell receptor alpha chain (TRAC) in Jurkat cells using CRISPR ( figure 1 ). The two specific guide sequences used for CD3z KO were (i) CAGTTGCCGATTACAGGTA and (ii) GTGGAAGGCGCTTTTCACCG. The resulting cells were analyzed by flow cytometry to show the effect of knockdown on the expression of the CD3 / T cell receptor complex. Both knockouts abolished the expression of the CD3 / T cell receptor in these cells compared to mock electroporated (EP) cells, and as determined by using anti-CD3e antibody at day 7 post EP. CD3ζ or TRAC ( figure 2 ). U...
Embodiment 2
[0248] Example 2: Modification of CD3ζ knockout T cells to reduce rejection
[0249] Construction of lentiviral vectors for use in transduction of T cells ( Figure 9 ). Nunchucks (pMGH81) is a gene expressing three full-length CMV proteins (UL40, US6, and UL18), click beetle green luciferase (CBG; to be used in cell killing assays and in vivo imaging), enhanced green fluorescent protein (EGFP; for downstream flow sorting), and viral 2A proteins (T2a, P2A, E2A, and F2A; for directing cleavage of the encoded polyprotein). As explained above, CMV proteins function to facilitate evasion of the immune attack of the recipient to whom T cells are administered.
[0250] Ninja (pMGH82) is a construct expressing a modified anti-human CD19_BBz chimeric antigen receptor, as well as CMV UL40CMV viral protein and signal peptide. A signal peptide is loaded onto non-classical HLA-E which, when expressed, helps inhibit NK cell killing. Also included are CMV US6 and UL18, and mCherry as ...
Embodiment 3
[0253] Embodiment 3: Knockout T cell receptor alpha chain (TRAC)
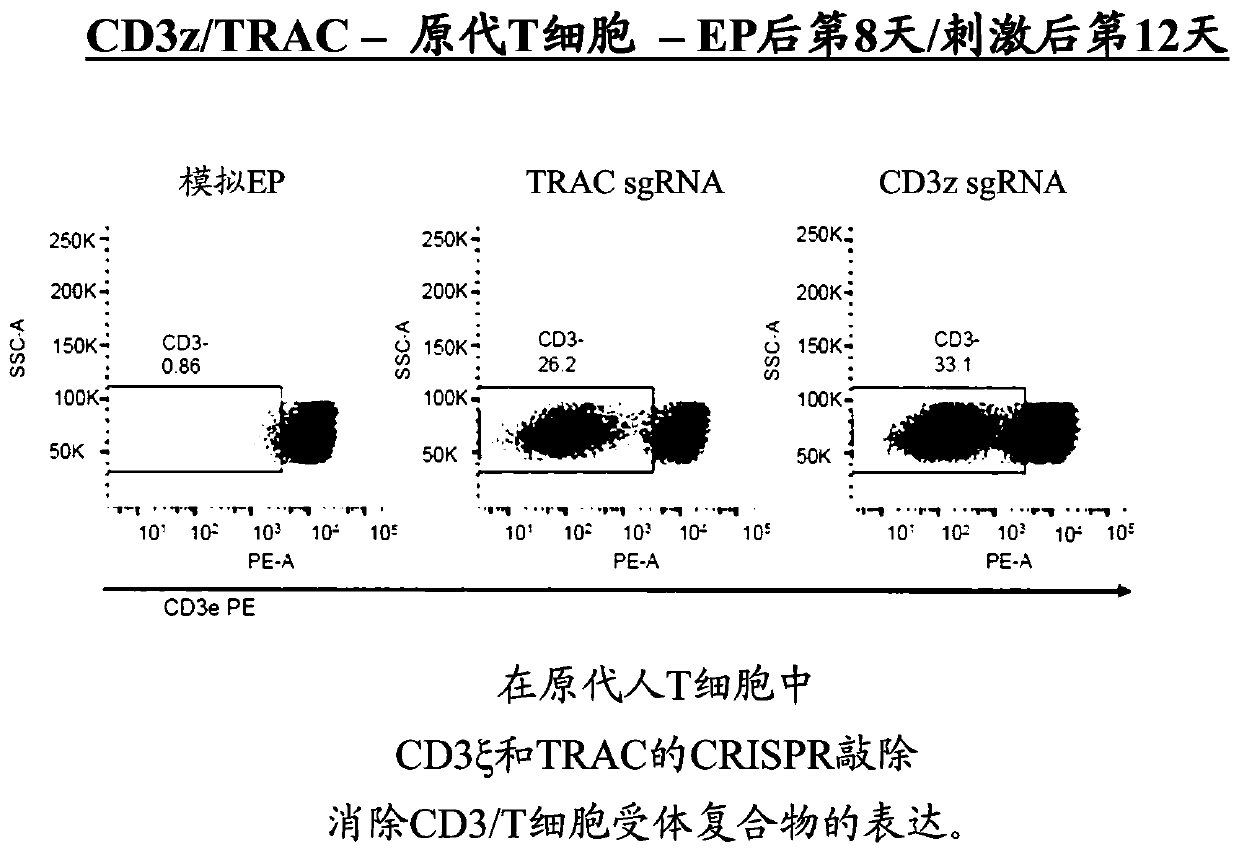
[0254] TRAC was targeted using an approach similar to that described above for CD3ζ. The specific gRNA used for TRAC is AGAGTCTCTCAGCTGGTACA. Such as Figure 14 As shown in , knockdown of TRAC (AGAGTCTCTCAGCTGGTACA) or CD3ζ (GTGGAAGGCGCTTTTCACCG) in primary T cells caused knockdown of cell surface expression of TCR at day 8 after electroporation / day 12 after stimulation.
[0255] Various aspects of the invention are described in the following numbered paragraphs.
[0256] 1. An isolated T lymphocyte modified to have reduced expression of CD3ζ, T cell receptor alpha chain (TRAC), and / or T cell receptor beta chain (TRBC) genes due to reduced or eliminated CD3ζ Depleted or abolished T cell receptor (TCR) expression.
[0257] 2. The isolated T lymphocyte of paragraph 1, which comprises a CD3ζ, TRAC, and / or TRBC gene, regulatory sequence, coding sequence, exon, or portion thereof mutated, resulting in reduced,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com