Construction of next generation sequencing (NGS) libraries using competitive strand displacement
A sequencing and dependency technology, applied in the field of NGS library construction, can solve problems, reduce reaction efficiency, consume flow cell space, etc., achieve excellent recovery, reduce the number of reads, and improve the detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
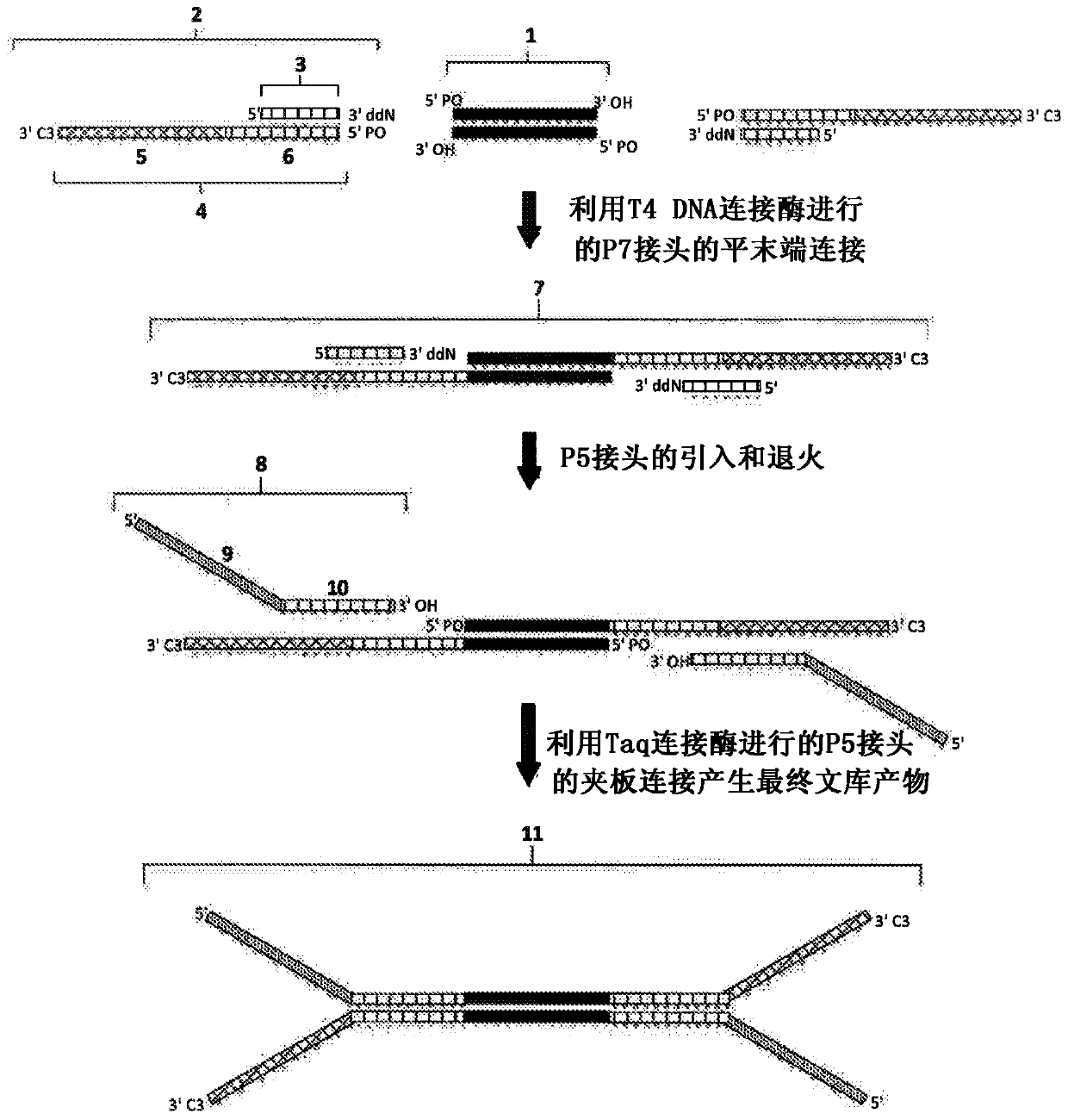
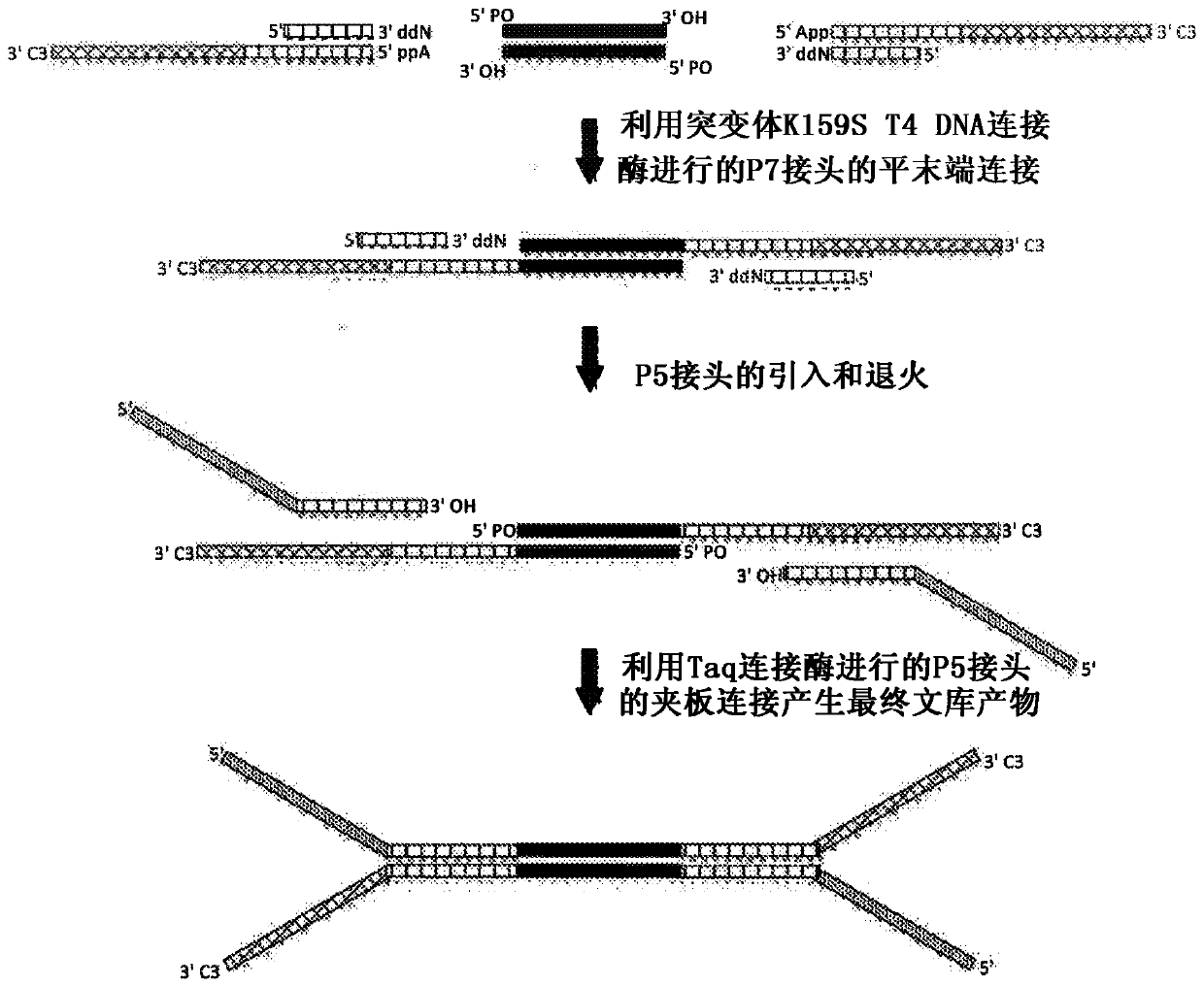
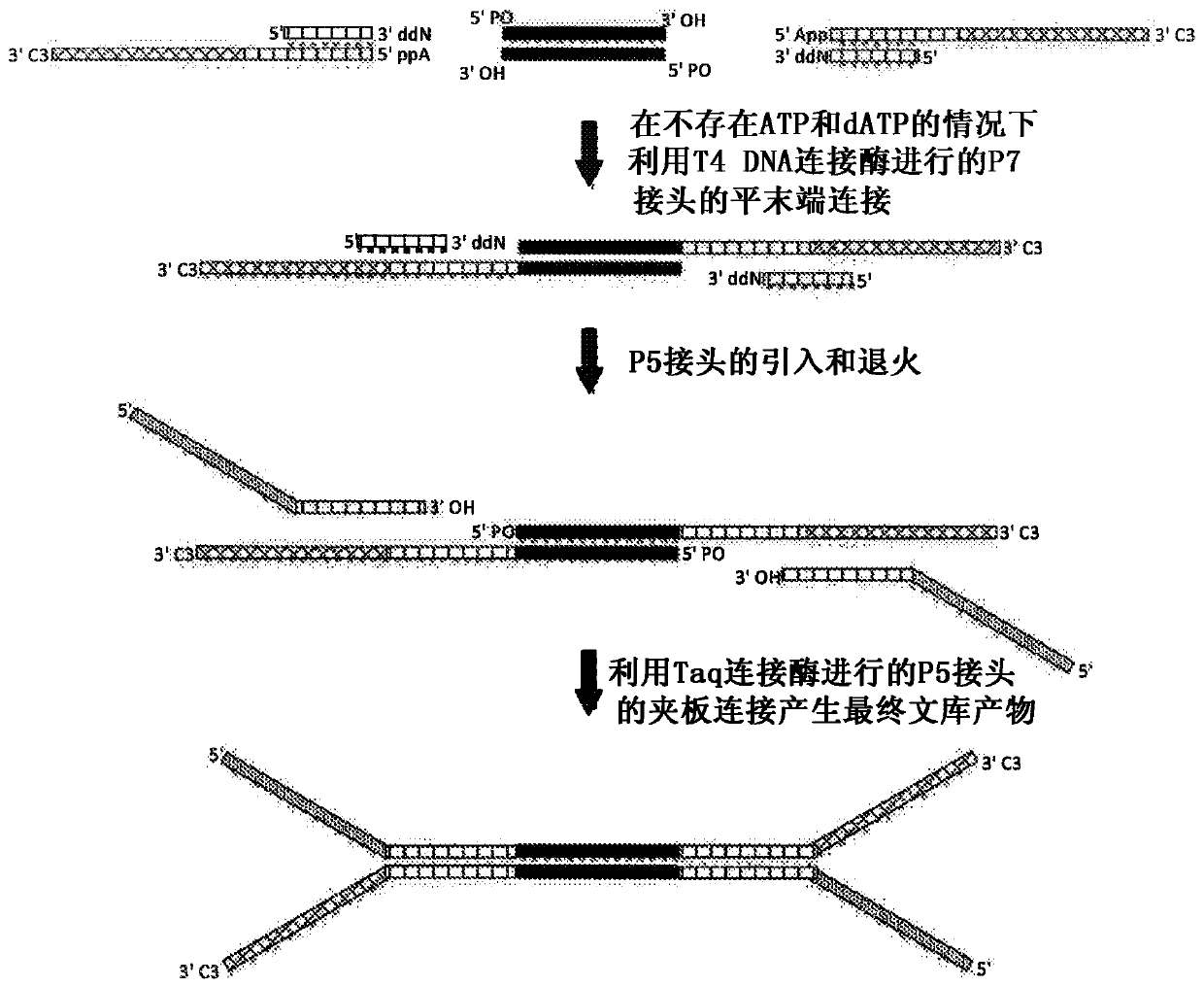
[0050] This example demonstrates that, with the use of Ultra TM II library (New England BioLabs) or KapaHyper Prep (Kapa Biosystems) method obtained enhanced coverage depth obtained from NGS libraries prepared from high quality genomic DNA using the second embodiment of the CSD method. High-quality genomic DNA was extracted from the cell line NA12878 (ATCC). 1 or 10 ng of extracted DNA was sheared to an average size of 150 bp using sonication (Covaris S220), followed by end repair, which included phosphorylation of the 5' end with T4 polynucleotide kinase (PNK) for 30 minutes, followed by purification by 2.5X AMPure beads. For CSD processing, a P7 linker (SEQ ID NO: 11-16) hybridized to a truncated 3'ddN-blocked oligonucleotide (SEQ ID NO: 17) was ligated by blunt end ligation using mutant K159S T4 DNA ligase Ligation to end-repaired target fragments for 15 minutes followed by a 15 minute heat kill step. A P5 adapter (SEQ ID NO: 1 or SEQ ID NO: 2) was then ligated to the...
Embodiment 2
[0052] This example demonstrates that, with the use of Ultra TMThe depth of coverage obtained from NGS libraries prepared from circulating cell-free DNA (cfDNA) using the second embodiment of the CSD method was enhanced compared to the depth of coverage obtained with the II library. The “real” cfDNA samples were real cell-free DNA isolated from healthy individuals by Biochain, while the “mock” cfDNA samples were cell line genomic DNA (NA12878) sheared to 150 bp using Covaris S2. Libraries were prepared in triplicate from 1 ng of cfDNA using the CSD and NEB methods as described in Example 1. When compared to the NEB method, the average coverage depth of CSD was 3.6 times higher with the "real" cfDNA input and 2.3 times higher with the "mock" cfDNA input ( Figure 8C ).
Embodiment 3
[0054] This example demonstrates the enhanced depth of coverage obtained using the second embodiment of the CSD method from NGS libraries prepared from low quality genomic DNA extracted from FFPE samples compared to the depth of coverage obtained using NEB Ultra II libraries. FFPE samples were purchased from Asterand Bioscience. Libraries were prepared using as starting material 1 ng, 5 ng, or 10 ng of FFPE-derived genomic DNA sheared to an average size of 200 bp, as described above. When compared to the NEB method, the average coverage depth of CSD was 1.8-fold, 1.4-fold, and 1.3-fold higher with 1 ng, 5 ng, or 10 ng FFPE-derived genomic DNA, respectively ( Figure 8D ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com