Genome unit point variation pathogenicity prediction method and system and storage medium
A prediction method and a single-point technology, applied in the field of bioinformatics, can solve the problems of high cost, low prediction accuracy, and low reliability, and achieve the effects of cost saving, convenient use of symptoms, and protection of patient privacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0030] The following will clearly and completely describe the technical solutions in the embodiments of the present invention in conjunction with the embodiments of the present invention and the accompanying drawings in the embodiments:
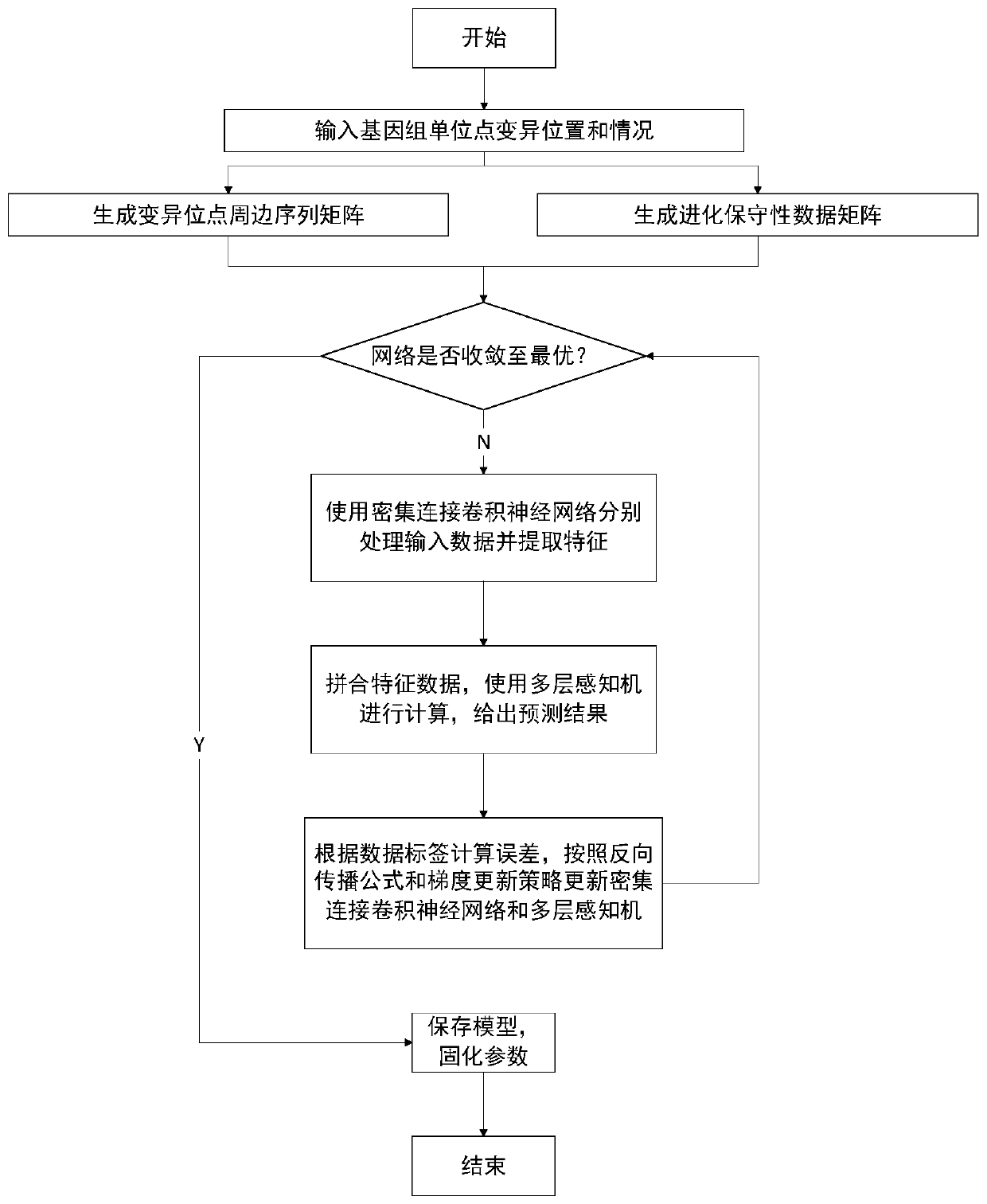
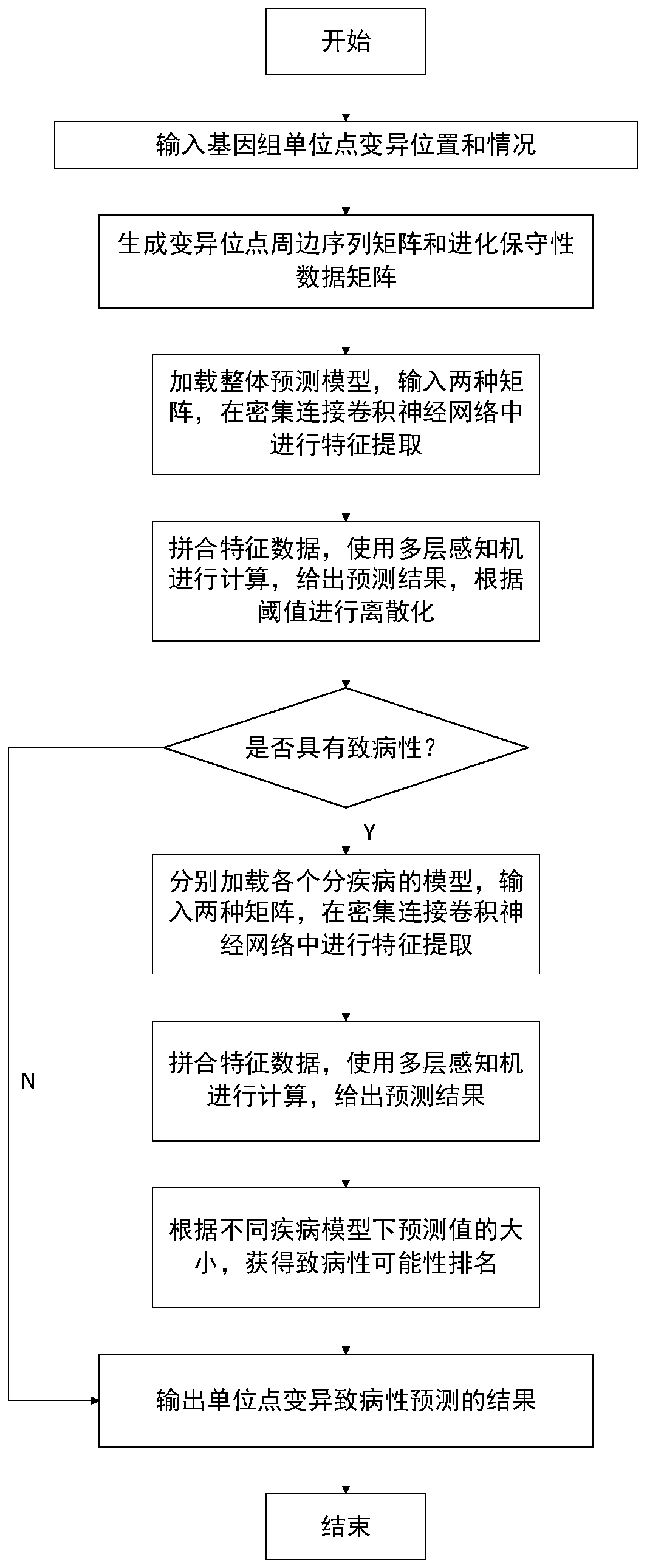
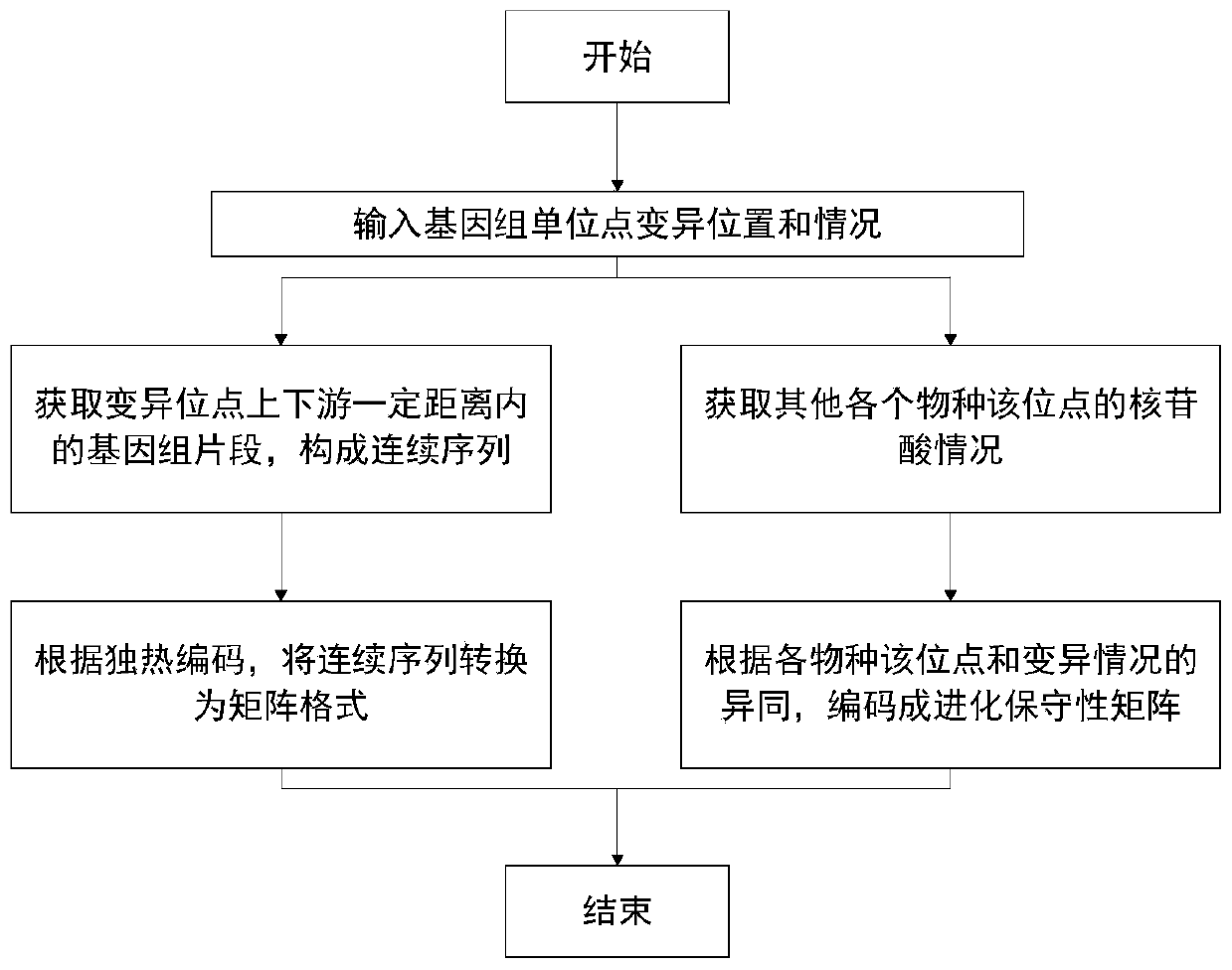
[0031] The method for predicting the pathogenicity of genome single-site mutations provided by the present invention is based on deep learning methods, using genome single-site mutation data obtained by sequencing and reference genome sequences (also known as evolutionary conservation data) of various species as training data, and using The medical diagnosis result is used as a category label, and the model obtained after training can calculate the data of various single-point variations and predict the probability of causing genetic diseases according to different needs.
[0032] The model basis of the present invention is a hybrid of convolutional neural networks and multi-layer perceptrons. The purpose of the convolutional neural network i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com