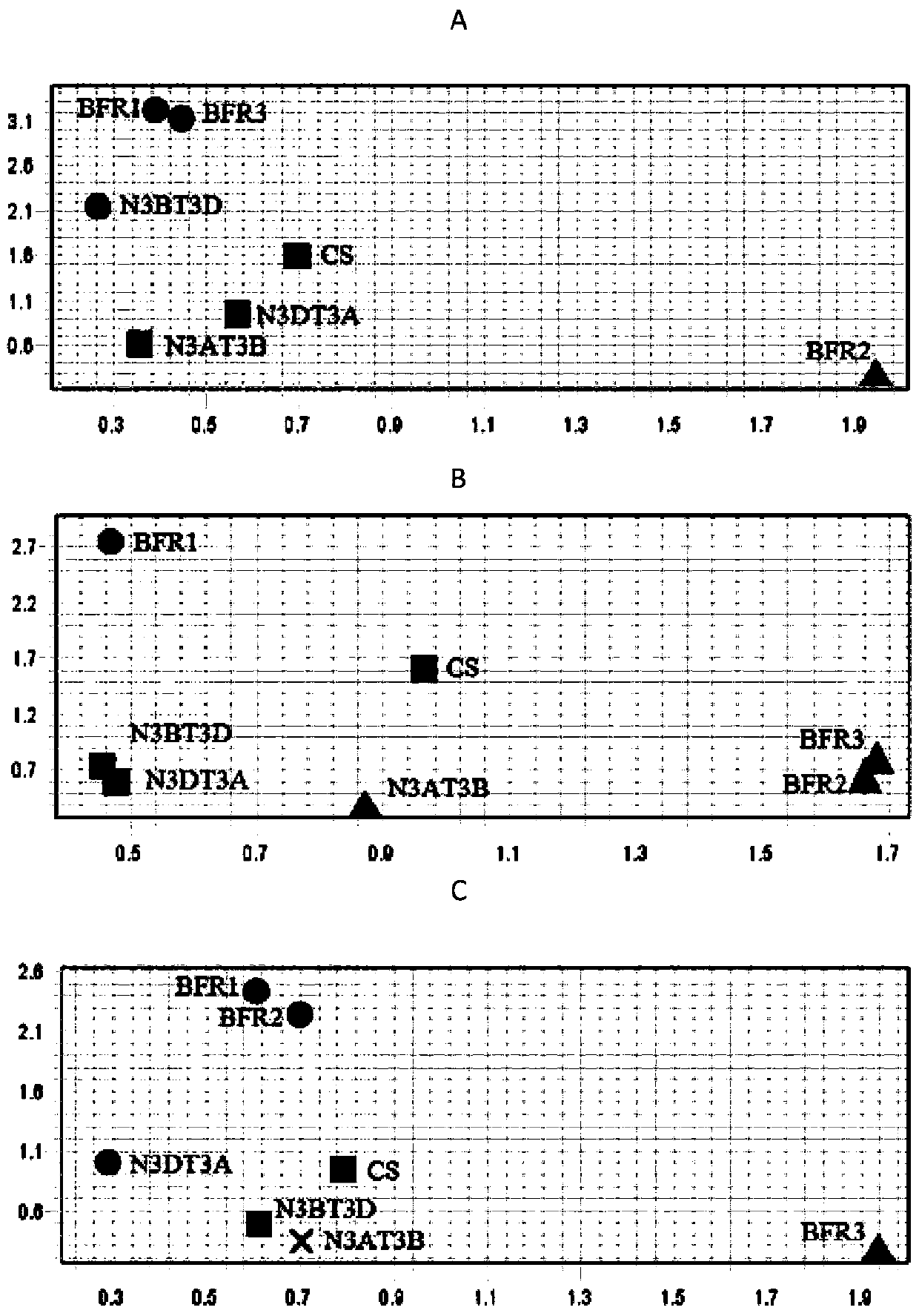
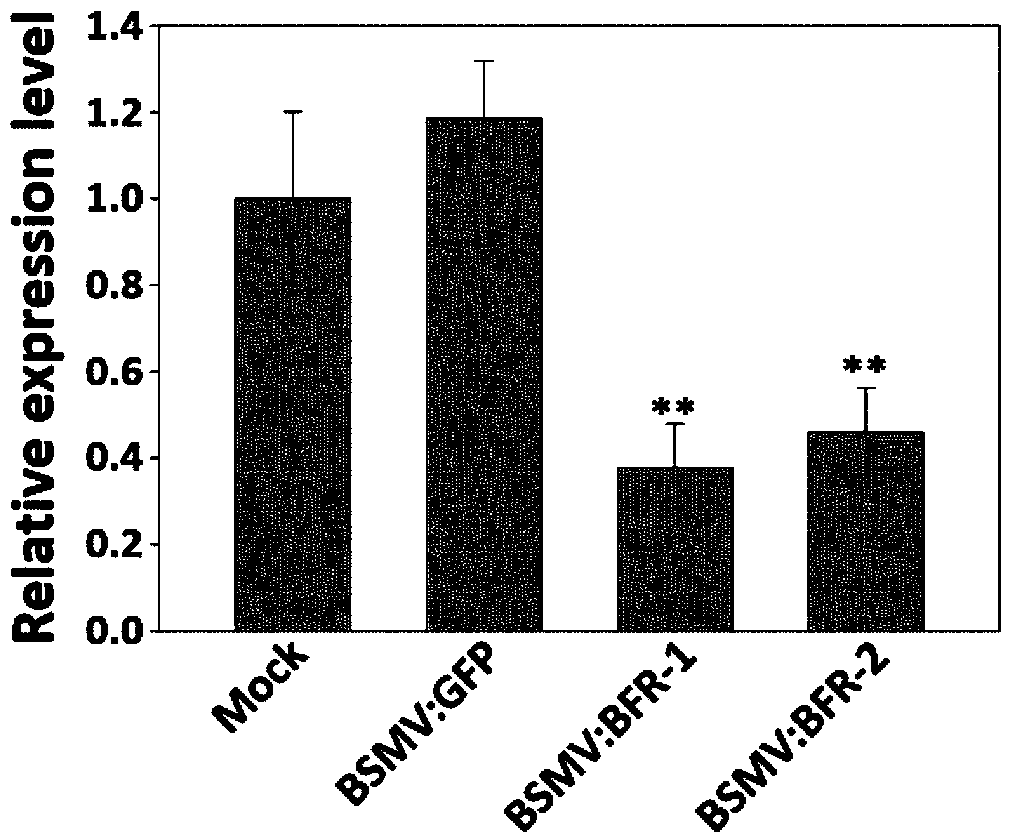
BFR protein associated with resistance of wheat to powdery mildew, and coding gene and application thereof
A powdery mildew and protein technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of reduced wheat quality, threats to wheat production and food security, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment one, the cloning of BFR gene cDNA and DNA sequence
[0034] 1. Acquisition of wheat cDNA and genome sequences
[0035] 1) Extraction of wheat genomic DNA and total RNA
[0036] Take 0.5 g of fresh wheat leaves, put them into a 2 mL centrifuge tube equipped with steel balls, freeze them with liquid nitrogen, and grind the materials into powder in a tissue grinder. Add 1.0mL lysate solution (1% SLS, 417mM Tris / HCl pH8.0, 417mM NaCl, 83mM EDTA), shake vigorously, vortex and mix thoroughly, and let stand on ice for 10min to fully lyse the cells. Add the extraction solution (Tris saturated phenol / chloroform / isoamyl alcohol = 25:24:1) equal to the volume of the lysate into the centrifuge tube, shake slowly until the emulsion is formed, let stand on ice (or room temperature) for 10 minutes, 12000rpm Centrifuge for 10 min, and transfer the supernatant to a new centrifuge tube. Repeat this operation once. Pipette the supernatant into another 1.5mL centrifuge tube,...
Embodiment 2
[0050] Example 2. Chromosomal mapping of the BFR gene using KASP markers
[0051] 1. Design and synthesis of KASP molecular marker primers
[0052] 1) The 3 BFR gene copies amplified from wheat genomic DNA in Example 1 were homologously compared, and 162 (A / G), 643 (G / A) and 1245 (C / T) were selected respectively Primers were designed for the SNP site at the bp position.
[0053]2) Design the selected SNP site and its upstream 19nt as two forward primers, and then add FAM: 5'-GAAGGTGACCAAGTTCATGCT-3' or VIC: 5'-GAAGGTCGGAGTCAACGGATT- at the 5' end of the two forward primers 3' fluorescent reporter tag sequence. Then select a suitable position downstream of the SNP site and select a nucleotide sequence of about 20 nt as a reverse primer, so that the length of the fragment obtained by the final PCR amplification is 50-80 bp. The reverse primer is not tagged. The KASP marker primers for the three SNP sites are:
[0054] SNP162F1: 5'-gaaggtgaccaagttcatgctCTGAGGGTCTTGTTGCTGCA-3...
Embodiment 3
[0071] Embodiment three, utilize BSMV-VIGS experiment to verify the anti-powdery mildew function of BFR gene
[0072] 1. Obtaining wheat with silenced BFR gene
[0073] 1) Induction of BFR gene down-regulation and construction of BSMV-VIGS vector system
[0074] (1) According to the BFR-3B gene sequence in Example 1, two silent fragments were selected in the non-conserved region outside the basic-Helix-Loop-Helix domain, and one fragment was located near the N-terminus of the coding region, the specific position It is +109bp~+314bp, 206bp long, named as the silent sequence BFR-1, using the primer pair V-BFR-F1: 5'-ATT GCTAGC TTTGCATACTTGACAAAAG-3' and V-BFR-R1:5'-TAA GCTAGC GGTTCACGGCAAGAGC-3' was obtained by PCR amplification. There is a SNP difference between this fragment and BFR-3A-xiaobaidongmai at the +161bp position, and there is a SNP difference between the BFR-3D-xiaobaidongmai at the +161bp and +203bp positions respectively. Another fragment is near the C-termi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com