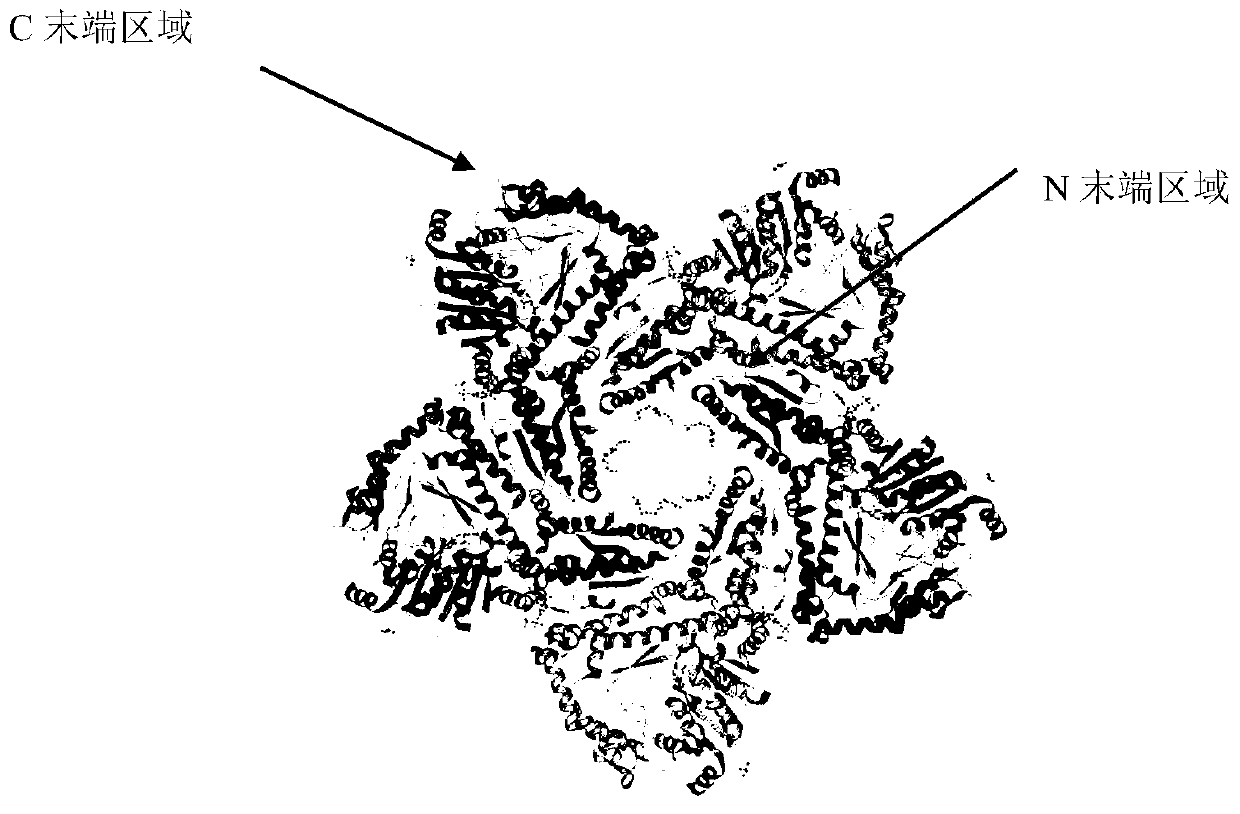
Molecular modification method for improving activity and stability of lysine decarboxylase
A technology of lysine decarboxylase and molecular transformation, which is applied in the field of molecular transformation to improve the activity and stability of lysine decarboxylase, which can solve the problems of poor stability and alkali resistance of lysine decarboxylase, and achieve the improvement of catalytic activity , avoid uncertainty and tedious screening process, good stability and catalytic activity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 1. Obtain the gene fragment encoding lysine decarboxylase molecule, gene sequence (SEQ ID NO.1, GenBank Accession number: NC_000913).
[0041] 2. Link the gene fragment (SEQ ID NO.2) encoding the EK polypeptide to the 5' end of the gene fragment encoding the lysine decarboxylase molecule (SEQ ID NO.1, GenBank Accession number: NC_000913). This embodiment uses PCR method, the specific operation method is as follows:
[0042] (1) Primer design: CadA-F-EK: SEQ ID NO.6; CadA-R: SEQ ID NO.7
[0043] (2) Using SEQ ID NO.1 as a template and using SEQ ID NO.6 and SEQ ID NO.7 as primers to perform a PCR reaction to obtain SEQ ID NO.3;
[0044] PCR cycling process (Taq enzyme):
[0045] Step1: 94°C for 3 minutes
[0046] Step2: 94℃30s
[0047] Step3: 55℃30s
[0048] Step4: 128s at 72°C (to step2, 30 cycles)
[0049] Step5: 5min at 72°C
[0050] Detection of PCR products: Take 5 μL of PCR products and detect them by agarose gel electrophoresis.
[0051] 3. Ligate the SEQ I...
Embodiment 2
[0054] 1. Obtain the gene fragment encoding lysine decarboxylase molecule, gene sequence (SEQ ID NO.1, GenBank Accession number: NC_000913).
[0055] 2. Link the gene fragment (SEQ ID NO.2) encoding the EK polypeptide to the 3' end of the gene fragment (SEQ ID NO.1, GenBank Accession number: NC_000913) encoding the lysine decarboxylase molecule. This embodiment uses PCR method, the specific operation method is as follows:
[0056] (1) Primer design: CadA-F: SEQ ID NO.8; CadA-R-EK: SEQ ID NO.9
[0057] (2) Using SEQ ID NO.1 as a template and using SEQ ID NO.8 and SEQ ID NO.9 as primers to perform a PCR reaction to obtain SEQ ID NO.4;
[0058] The PCR cycle process is the same as in Example 1.
[0059] 3. Connect the SEQ ID NO.4 obtained in step 2 to the pET-32a(+) vector, and transform it into the host Escherichia coli BL21(ED3), the specific operation method is as follows:
[0060] Use Bam HI and Hind III to double-digest plasmid pET-32a(+) and SEQ ID NO.4, and use T4 DNA...
Embodiment 3
[0062] 1. Obtain the gene fragment encoding lysine decarboxylase molecule, gene sequence (SEQ ID NO.1, GenBank Accession number: NC_000913).
[0063] 2. The gene fragment (SEQ ID NO.2) encoding the EK polypeptide is simultaneously connected to the 5' end and the 3' end of the gene fragment encoding the lysine decarboxylase molecule (SEQ ID NO.1, GenBank Accession number: NC_000913), This embodiment adopts PCR method, and concrete operation method is as follows:
[0064] (1) Primer design: CadA-F-EK: SEQ ID NO.6; CadA-R-EK: SEQ ID NO.9
[0065] (2) Using SEQ ID NO.1 as a template and using SEQ ID NO.6 and SEQ ID NO.9 as primers to perform a PCR reaction to obtain SEQ ID NO.5;
[0066] The PCR cycle process is the same as in Example 1.
[0067] 3. Link the SEQ ID NO.5 obtained in step 2 to the pET-32a(+) vector, and transform it into the host Escherichia coli BL21(ED3), the specific operation method is as follows:
[0068] Use Bam HI and Hind III to double digest plasmid pET-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com