Mutants of penicillin G acylase from Achromobacter sp. CCM 4824 and uses thereof
A technology of penicillin and acylase, which is applied to the penicillin G acylase mutant derived from Achromobacter sp.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
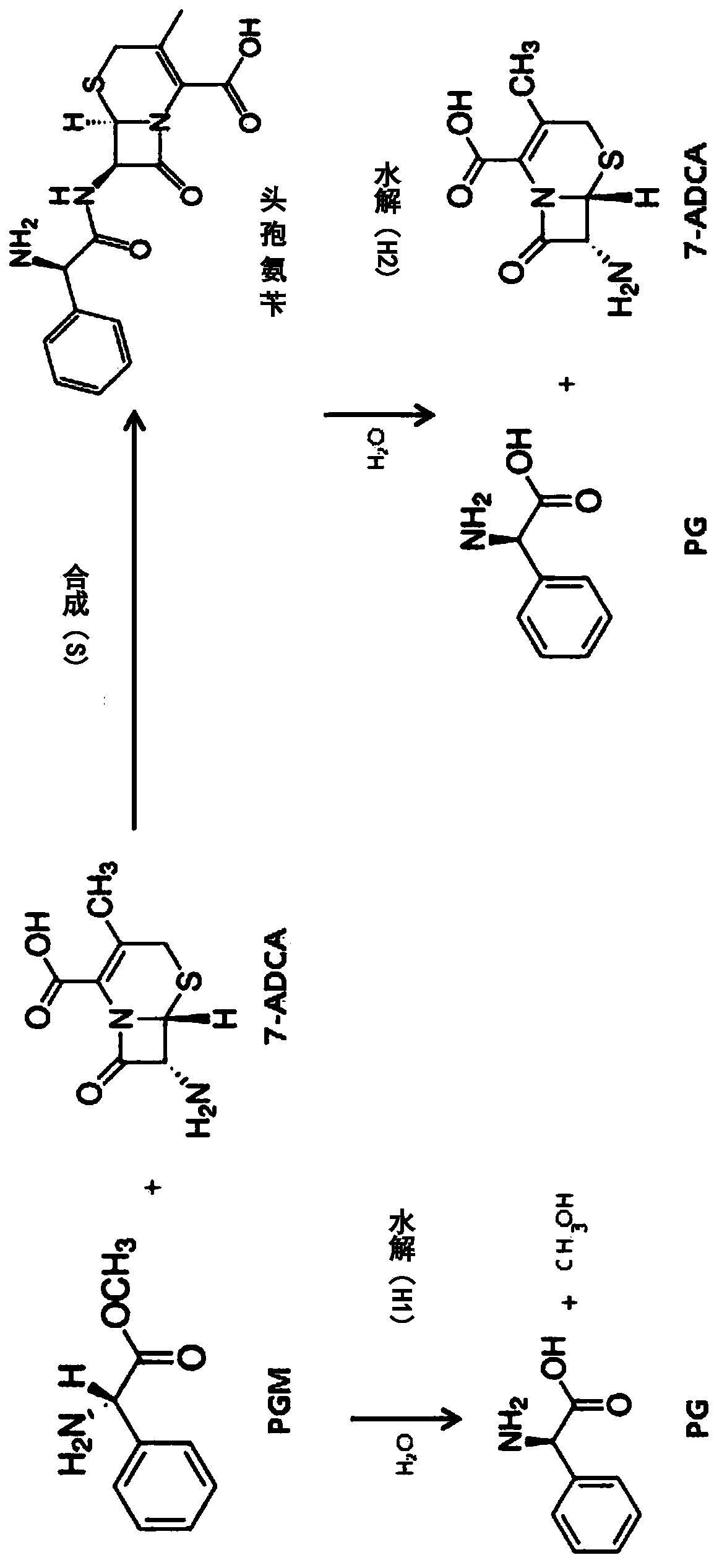
[0107] Preparation of wild-type Penicillium derived from Achromobacter sp. CCM 4824 Penicillin G acylase (APA)
[0108] Synthetic expression of wild-type penicillin G acylase gene derived from Achromobacter sp. CCM 4824
[0109] Wild-type APA (penicillin G acylase derived from Achromobacter sp. CCM 4824) is a precursor (precursor) type, through the α subunit shown in SEQ ID NO.1 and SEQ ID NO.2 The shown β subunit is expressed by a single-chain polypeptide (SEQ ID NO.4) connected and constituted by a spacer peptide shown in SEQID NO.3, and then through autocatalytic processing in cells to A mature active dimer form consisting of the α and β subunits is formed. The gene expressing wild-type APA precursor SEQ ID NO.4 (abbreviated as "apa gene") was synthesized by Bioneer Company (Daejeon, Korea).
[0110] Preparation of recombinant vector (pBC-APA) expressing wild-type APA
[0111] The apa gene obtained in was inserted into the XbaI and NotI restriction enzyme recogni...
Embodiment 2
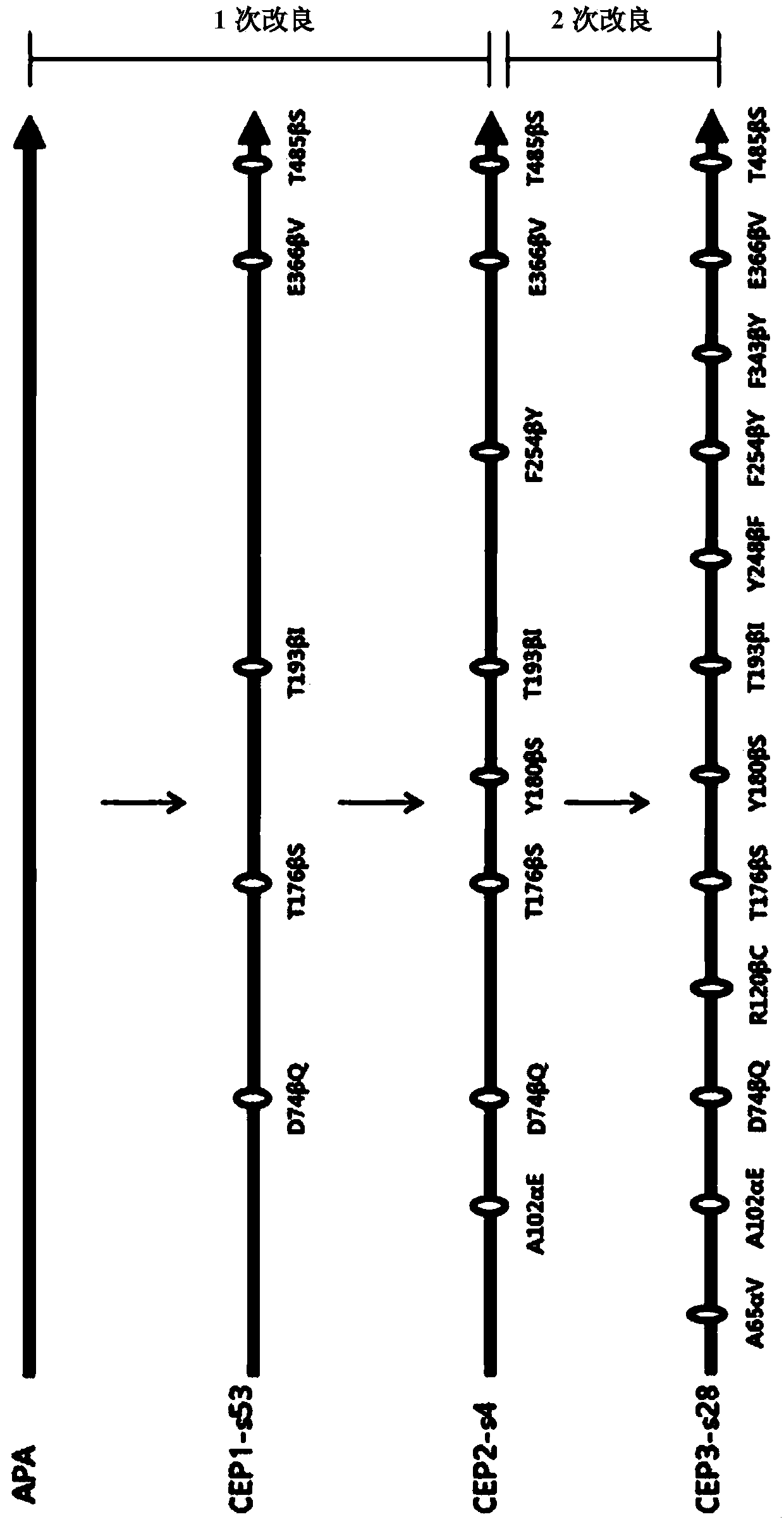
[0113] Generation and screening of mutations with high levels of multiplex synthesis performance against multiple β-lactam antibiotics body
[0114] Prepare mutant library once by error prone PCR
[0115] The nucleotide sequence of the apa gene synthesized in the was artificially randomly mutagenized, for which error prone PCR was performed to prepare a mutant library. The procedure for preparing a library of specific mutations is as follows.
[0116] Specifically, error-prone polymerase chain reaction was performed using the Diversity PCR Random Mutagenesis Kit (Diversity PCR Random Mutagenesis kit, Clontech, USA) to generate one mutation per 1,000 bp. PCR reaction solution, by using the template DNA of the pBC-APA plasmid of 1ng / μL, each 10pmol of T3 primer (SEQ ID NO.13) and T7 primer (SEQ IDNO.14), 2mM dGTP, 50X diversity dNTP mixture, 10X Titanium Taq buffer solution and titanium Taq polymerase, the final volume is 50 μL. The PCR reaction conditions were that th...
Embodiment 3
[0134] Preparation and screening of highly efficient antibiotic multiplex synthesis mutants with reduced substrate-decomposing activity
[0135] In order to prepare mutants having a high level of synthetic performance against various β-lactam antibiotics and significantly reduced hydrolytic activity of their reaction substrates, the following procedure was performed.
[0136] Prepare 5 times mutant library by error prone PCR
[0137] In order to increase the antibiotic (represented by cephalosporin) synthetic activity of the CEP2-s4 mutant prepared in the embodiment , and reduce the hydrolysis activity to the reaction substrate (represented by PGM), use CEP2- The s4 plasmid was used as template DNA, and an error-induced mutation library was prepared in the same manner as in Example .
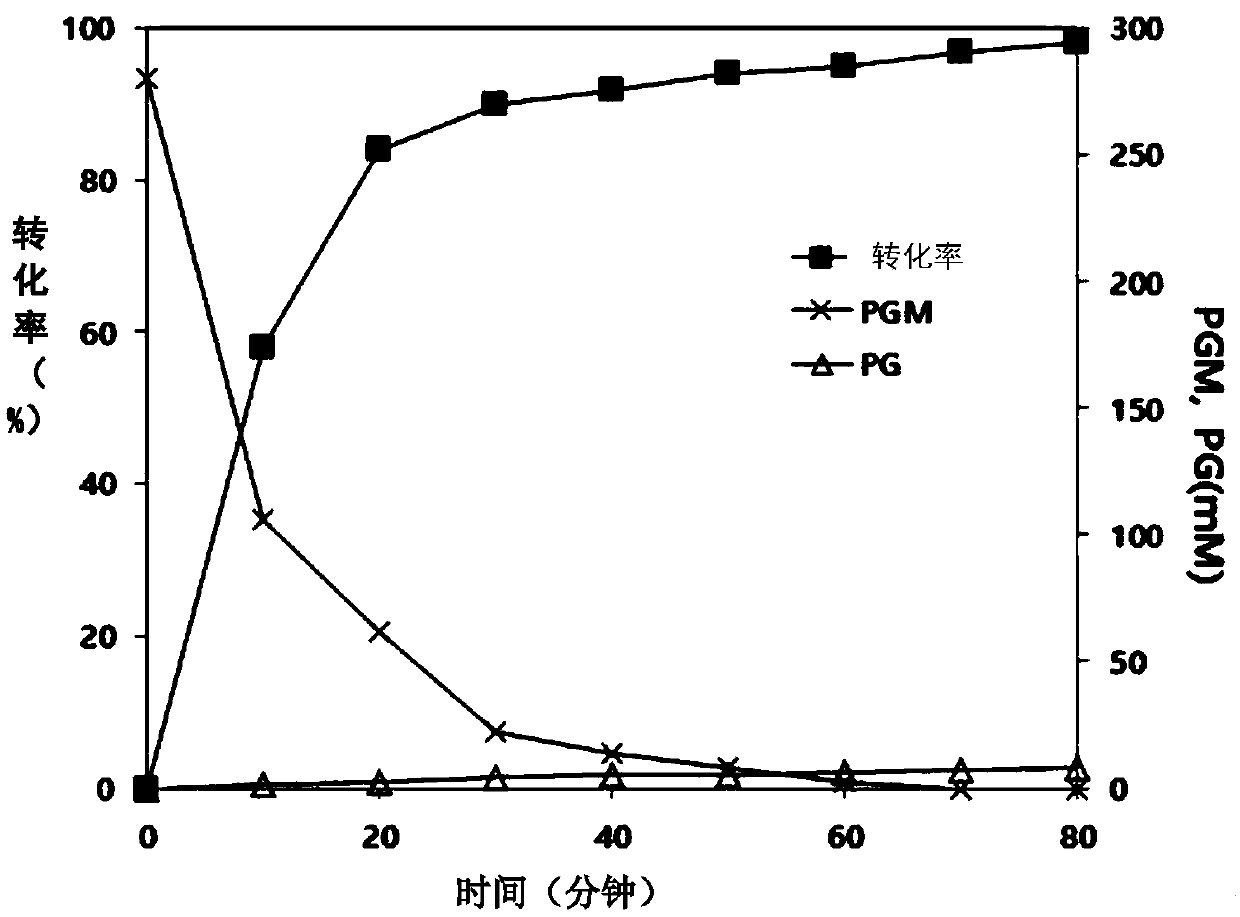
[0138] In order to study the synthetic ability to cephalosporins representatively, the test was carried out in the same manner as in Example . In addition, in order to study the reactivity...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com