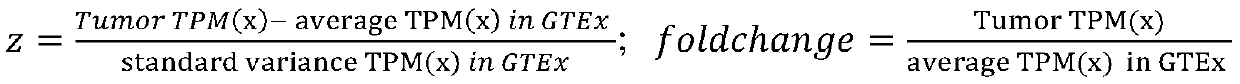Tumor antigen prediction method based on whole transcriptome, and application of the same
A technology of tumor antigen and prediction method, applied in the field of bioinformatics, which can solve the problems of incomplete assessment of tumor antigen load, not necessarily expressed mutations, incomplete mutation detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
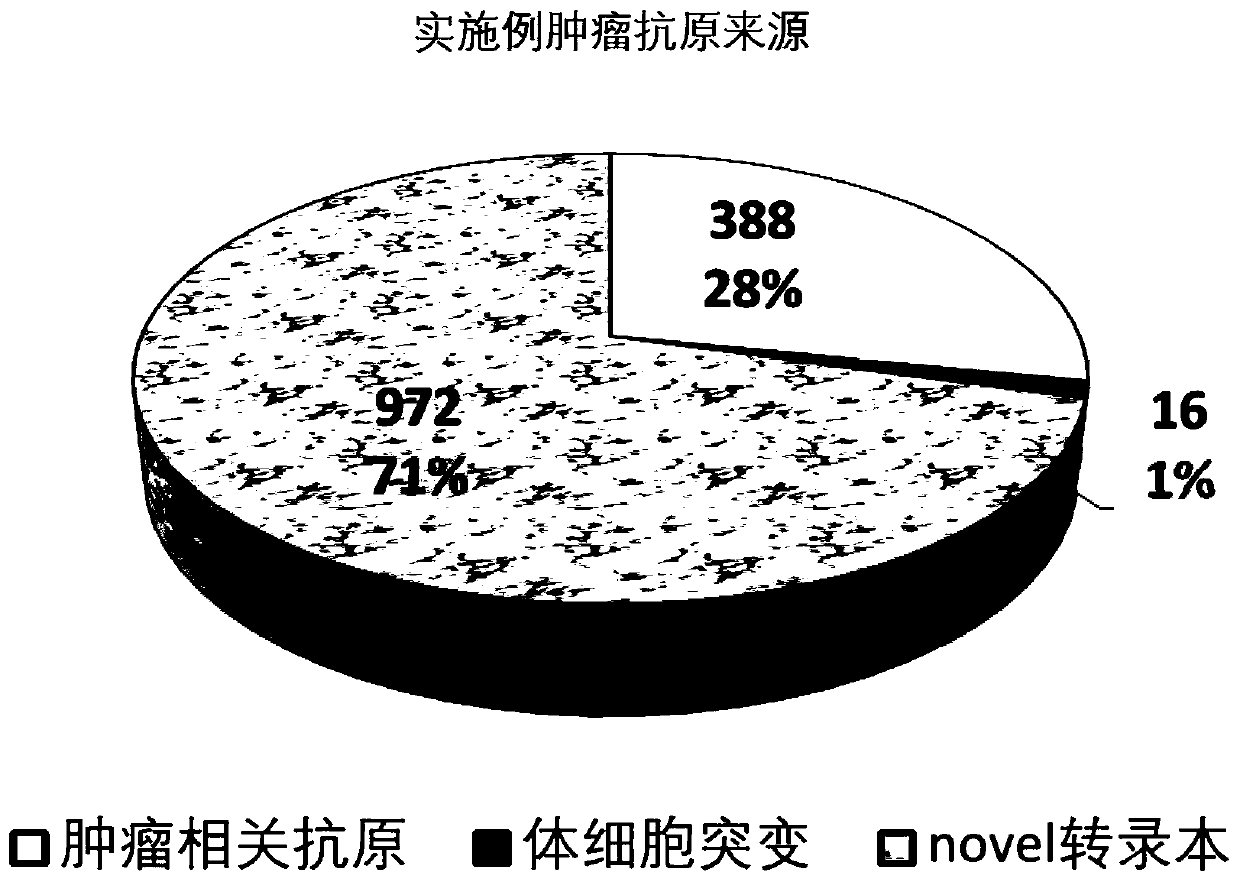
[0145] In this example, a pair of non-small cell lung cancer samples' tumor-adjacent whole-transcriptome sequencing data was used as input, and a total of 15 tumor-associated antigen genes, 3 tumor-specific gene fusions, and 488 tumors with protein coding capabilities were detected. novel transcripts and 127 somatic mutations in protein coding regions.
[0146] Table 1, Table 2, Table 3, and Table 4 respectively show the detailed information of tumor-associated antigen genes, gene fusions, novel transcripts, and somatic mutations. Due to space limitations, only the top20 are taken as examples.
[0147] Table 5 lists the detection results of four different HLA molecular typing tools Seq2HLA, OptiType, arcasHLA and hla-genotyper, A*31:01, B*40:01, C*07:02 have high consistency Allele was used for tumor antigen affinity prediction for this sample. Taking the affinity lower than 500nM as the cutoff, a total of 1376 tumor antigens were screened, that is, the overall antigen load o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com