A method for extract genomic DNA of Lonicera japonica Maxim
An extraction method and genome technology, applied in the field of molecular biology, can solve problems such as DNA browning
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
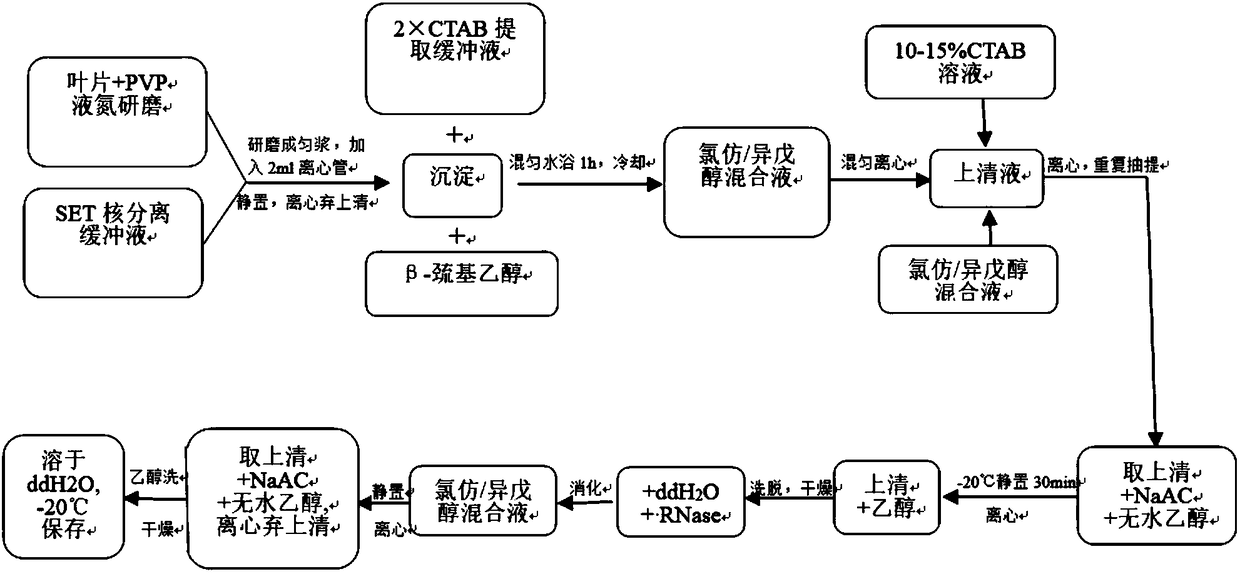
[0026] (1) Take 0.1 g of the leaves in a pre-cooled mortar, add a small amount of PVP powder, and immediately add 0.7-1.2 mL of SET nuclei separation buffer after liquid nitrogen grinding, quickly grind to a homogenate, and carefully transfer to a 2 mL centrifuge tube. Let stand at 2-8°C for 10min, centrifuge at 4000r / min for 10min at 4°C, and discard the supernatant.
[0027] (2) Add 700-1000μl 2×CTAB extraction buffer preheated at 60-65℃ for precipitation, then add 1-10μL of β-mercaptoethanol, mix well, put in water bath at 60-65℃ for 1 hour, shake gently several times during the period .
[0028] (3) After cooling to room temperature in a water bath, add 700-1000 μL of chloroform / isoamyl alcohol (22:3), gently invert to mix, and centrifuge at 12000 r / min for 10 min at room temperature.
[0029] (4) Take the supernatant to another tube, add 1 / 8-1 / 10 volume of preheated 10-15% CTAB solution and mix well, then add an equal volume of chloroform / isoamyl alcohol (22:3), lightly ...
example 2
[0035] (1) Take 0.1-0.35g leaves in a pre-cooled mortar, add a small amount of PVP powder, immediately add 0.7-1.2mL SET nuclei separation buffer after grinding with liquid nitrogen, quickly grind into a homogenate, and carefully transfer to a 2mL centrifuge tube . Let stand at 2-8°C for 10min, centrifuge at 4000r / min for 10min at 4°C, and discard the supernatant.
[0036] (2) Add 700-1000 μL of 2×CTAB extraction buffer preheated at 60-65°C to the precipitate, then add 1-10 μL of β-mercaptoethanol, mix well, and bathe in water at 60-65°C for 1 hour. During this period, shake gently for several times. Second-rate.
[0037] (3) After cooling to room temperature in a water bath, add 700-1000 μL of chloroform / isoamyl alcohol (23:2), mix gently by inversion, and centrifuge at room temperature at 12000 r / min for 10 min.
[0038] (4) Take the supernatant to another tube, add 1 / 8-1 / 10 volume of preheated 10-15% CTAB solution and mix well, then add an equal volume of chloroform / isoam...
example 3
[0044] (1) Take 0.1-0.35g of leaves in a pre-cooled mortar, add a small amount of PVP powder, and immediately add 0.7-1.2mL of SET nucleus separation buffer after liquid nitrogen grinding, quickly grind into a homogenate, and carefully transfer to a 2mL centrifuge tube middle. Let stand at 2-8°C for 10min, centrifuge at 4000r / min for 10min at 4°C, and discard the supernatant.
[0045] (2) Add 700-1000 μL of 2×CTAB extraction buffer preheated at 60-65°C to the precipitate, then add 1-10 μL of β-mercaptoethanol, mix well, and bathe in water at 60-65°C for 1 hour. During this period, shake gently for several times. Second-rate.
[0046] (3) After cooling to room temperature in a water bath, add 700-1000 μL of chloroform / isoamyl alcohol (24:1), mix gently by inversion, and centrifuge at room temperature at 12000 r / min for 10 min.
[0047] (4) Take the supernatant to another tube, add 1 / 8-1 / 10 volume of preheated 10-15% CTAB solution and mix well, then add an equal volume of chlo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com