Nitrogen efficient fusion gene SA and application thereof
A gene fusion and high-efficiency technology, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problems of excessive nitrogen fertilizer application and low nitrogen use efficiency, and achieve the effects of promoting vegetative growth, improving remobilization efficiency, and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
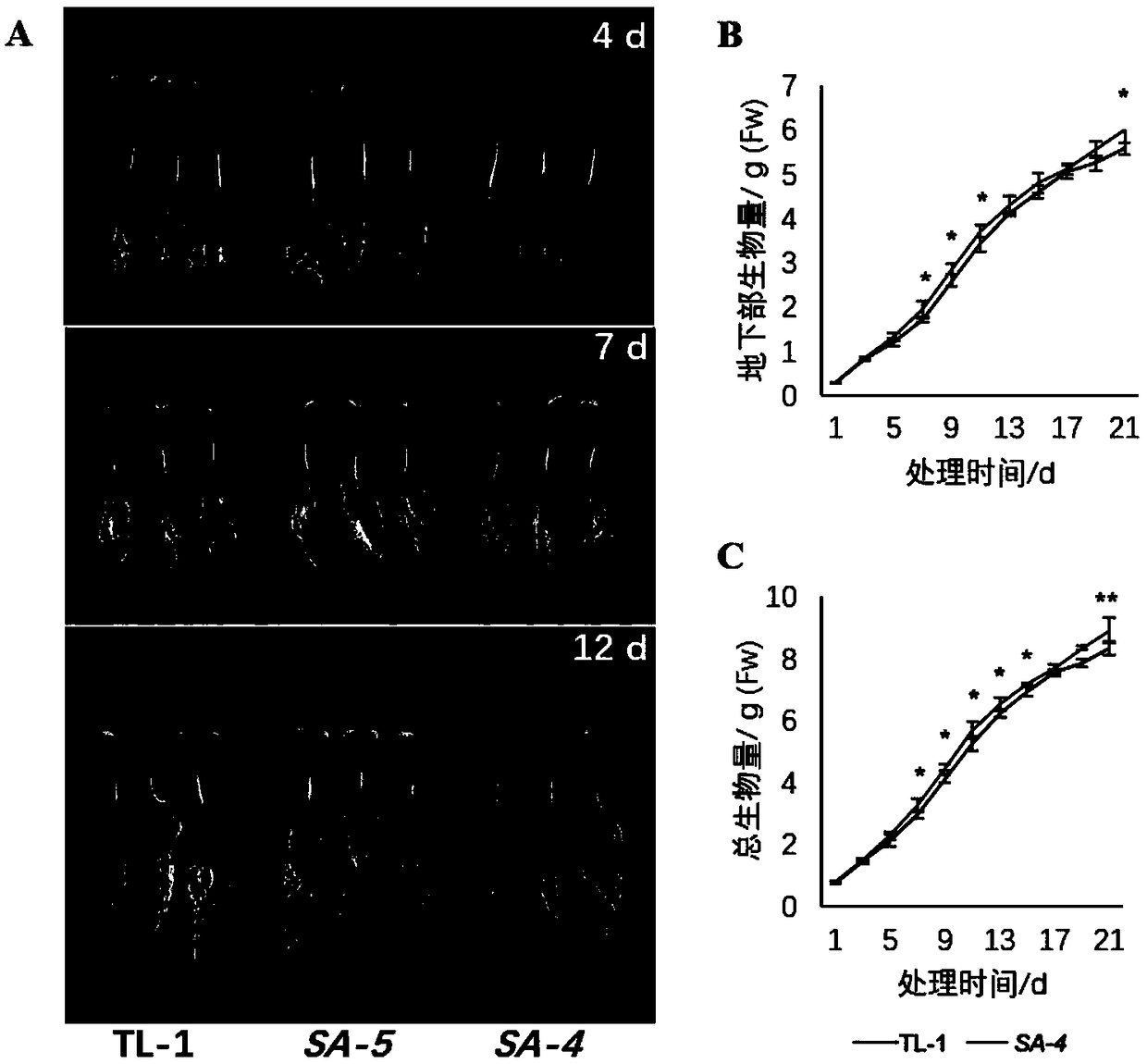
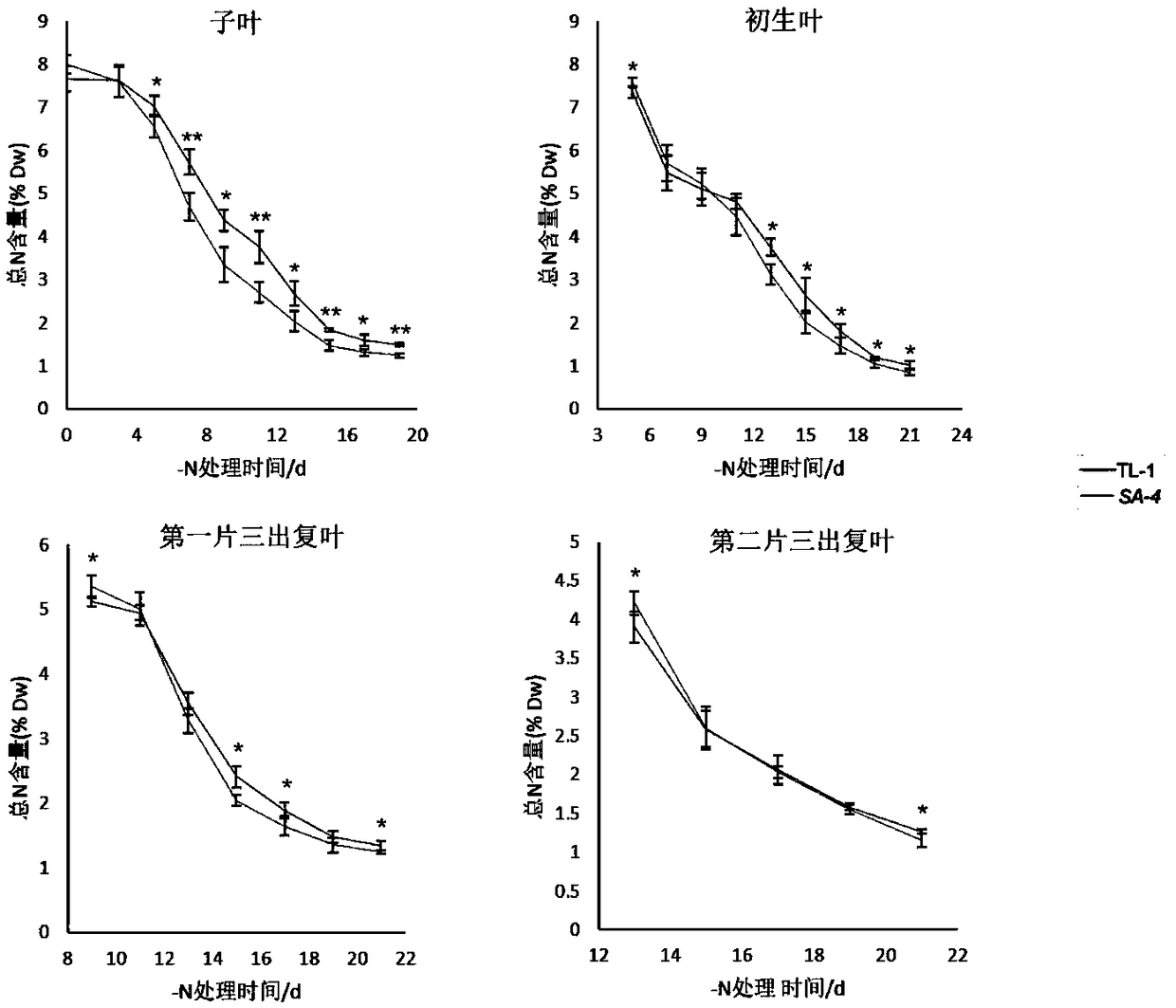
Image
Examples
Embodiment 1
[0052] Example 1: Cloning of Soybean "Origin" Organ-Specific Promoters
[0053] Using soybean genomic DNA as a template, the 1800bp promoter was amplified by PCR method, and the amplified product was recovered and cloned by TA.
[0054] (1) PCR amplification of the target fragment
[0055] Specific primers were designed according to the soybean "source" organ-specific promoter sequence, the sequences of which are shown in SEQ ID No.2 and SEQ ID No.3, wherein the downstream primer introduces the Nco I cutting point.
[0056] Genomic DNA of soybean was extracted according to CTAB method, and the genomic DNA was used as a template to carry out PCR amplification with the above primers to prepare gene promoter fragments.
[0057] PCR reaction system:
[0058]
[0059] PCR reaction program:
[0060] 94°C for 5 minutes;
[0061] 94°C for 30 seconds, 58°C for 110 seconds, 25 cycles;
[0062] 72°C for 10 minutes;
[0063] (2) Cloning of target fragments and identification of p...
Embodiment 2
[0073] Example 2: Cloning of soybean autophagy key genes
[0074] First, using soybean cDNA as a template, the PCR method was used to amplify a 360bp functional gene, and the amplified product was recovered and TA cloned.
[0075] (1) PCR amplification of the target fragment
[0076] Design specific primers according to the sequence of known key genes of soybean autophagy, the sequences are as shown in SEQ ID No.4 and SEQID No.5, Nco I restriction site is introduced in the upstream primer, BstP I enzyme is introduced in the downstream primer cut site.
[0077] The quasi-soybean RNA was extracted by the Trizol method and reverse-transcribed into cDNA. Using the cDNA as a template, PCR amplification was carried out using the above primers to prepare gene fragments.
[0078] PCR reaction system:
[0079]
[0080] PCR reaction program:
[0081] 94°C for 5 minutes;
[0082] 94°C for 30 seconds, 58°C for 30 seconds, 25 cycles;
[0083] 72°C for 10 minutes;
[0084] (2) Clo...
Embodiment 3
[0094] Embodiment 3: utilize pCAMBIA3301 carrier to construct SA fusion gene
[0095] (1) The vector plasmid pCAMBIA3301 (purchased from a reagent company) was extracted from corresponding Escherichia coli engineering bacteria, and the large vector fragment was recovered by double digestion with Hind III / BstP I.
[0096] (2) Extract the plasmid from the TA clone prepared in Example 1, digest it with Hind III / Nco I, and recover the promoter fragment by agarose gel electrophoresis.
[0097] (3) Extract the plasmid from the TA clone prepared in Example 2, digest it with Nco I / BstP I, and recover the functional gene fragment by agarose gel electrophoresis.
[0098] (4) The above three fragments were ligated overnight at 16° C. under the catalysis of ligase to complete the construction of the expression vector SA-pCAMBIA3301.
[0099]
[0100] (5) Transform Escherichia coli DH5α competent cells with the ligation mixture, the method is the same as in Example 1.
[0101] (6) selec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com