Method for detecting expansion of short tandem repeat
A short tandem repeat and sequence technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of sensitivity loss, low sequence insertion sensitivity, high sequence error rate, and achieve the effect of improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
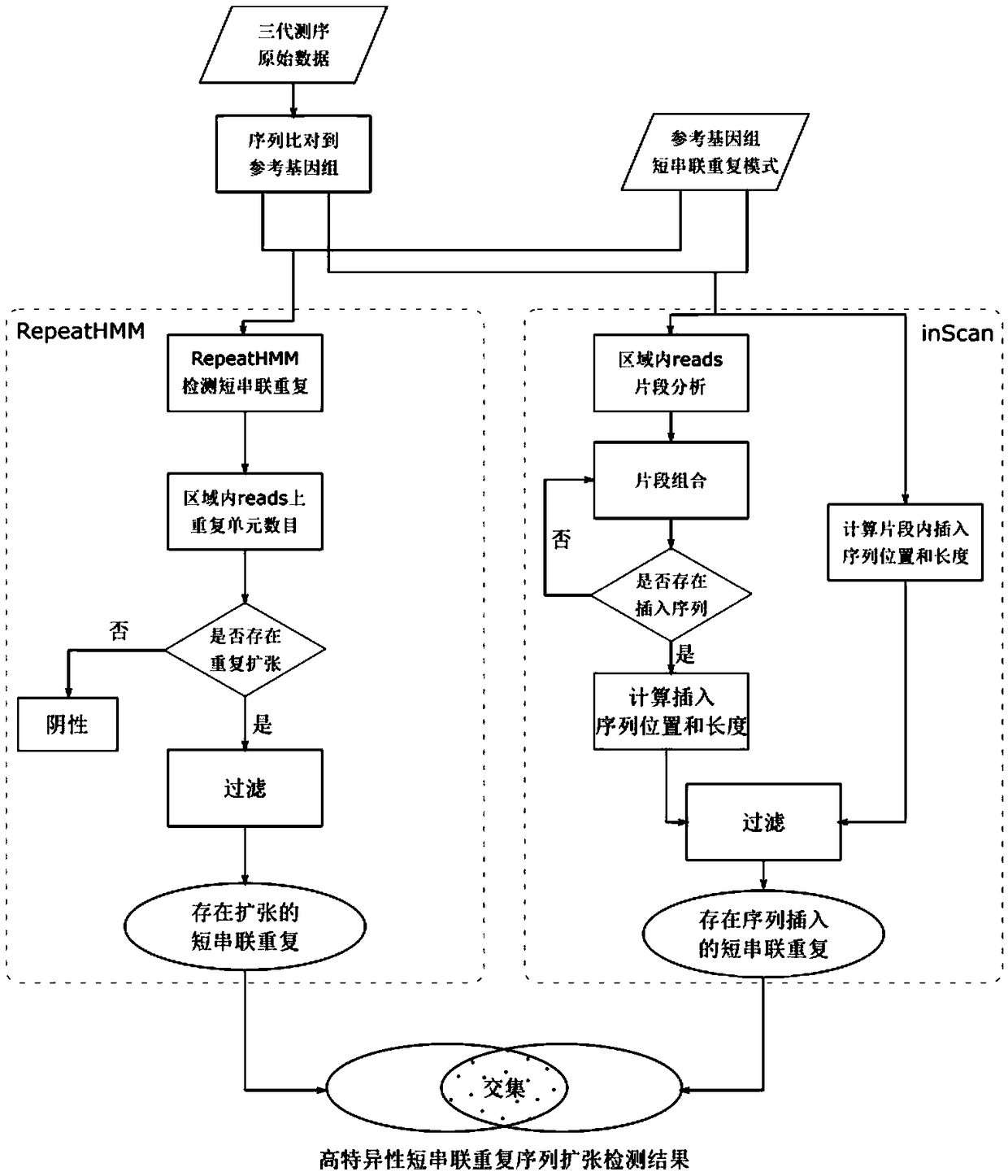
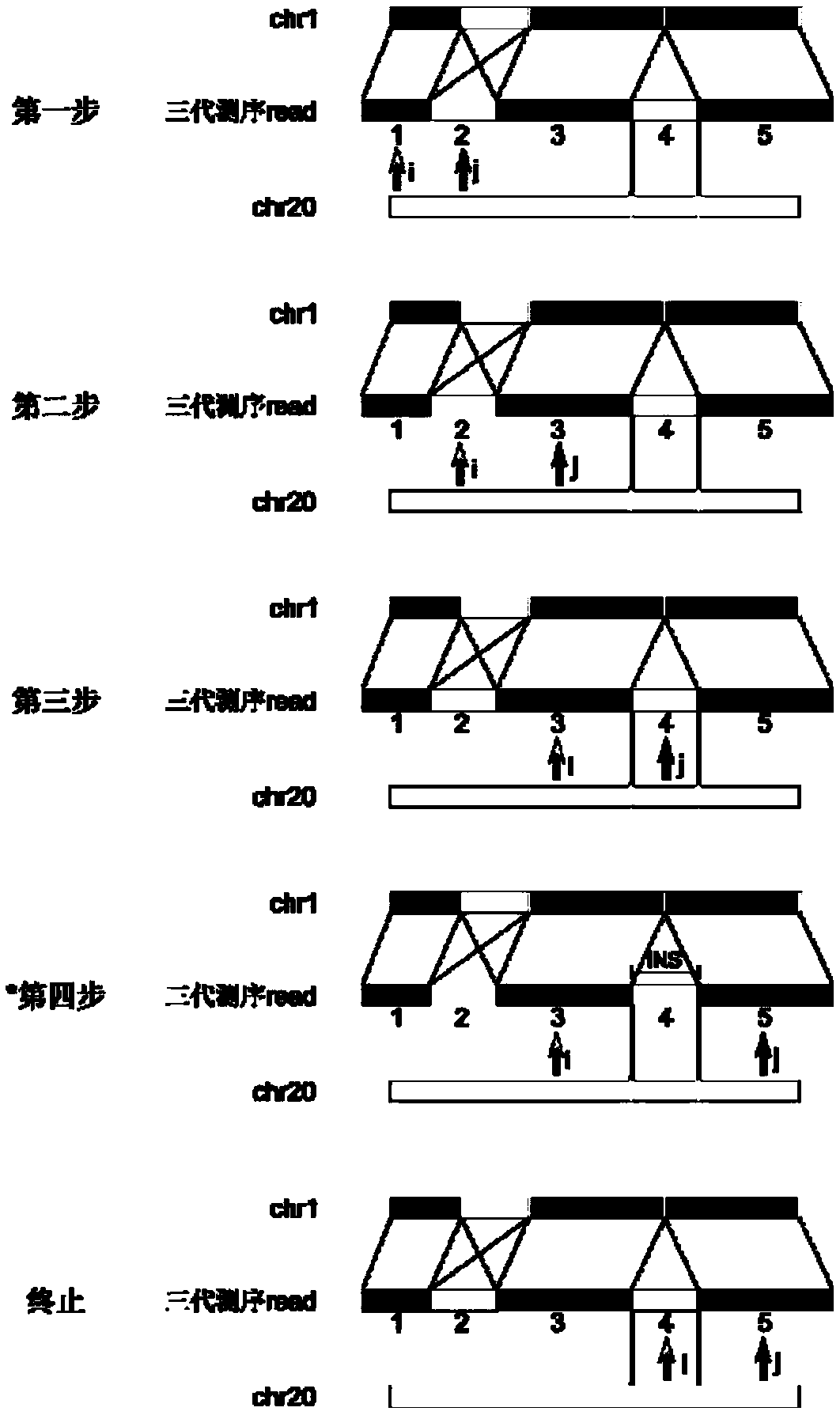
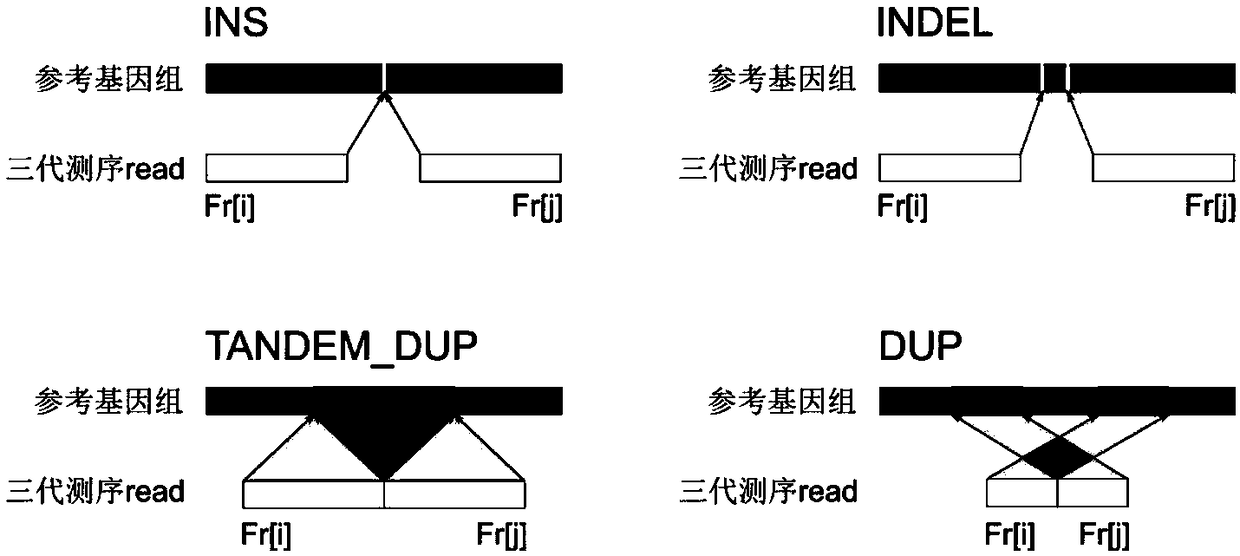
[0059] The next-generation sequencing data has a short read length (100-300bp), but the sequencing accuracy is high (>99%), and is suitable for detecting repeat expansions whose short repeat expansion length is smaller than the read length. In this example, HipSTR (Willems, T. et al. Genome-wide profiling of heritable and de novo STR variations. Nature Methods 14, 590 (2017)) was first used to detect NA12878 next-generation sequencing data (illuminaHiSeq2500, 150bp paired-end, 30X) The number of short tandem repeats in , randomly selected 1000 regions with short tandem repeat expansions and 4000 regions without short tandem repeat expansions as the test data set, and then used the test data set to test different detection methods to detect the third-generation sequencing data of NA12878 ( Pacbio Sequel sequencing platform, about 44X) Effect of short tandem repeat expansion. The main method flow chart of this technical solution is as follows: figure 1 Shown:
[0060] 1. Build...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com