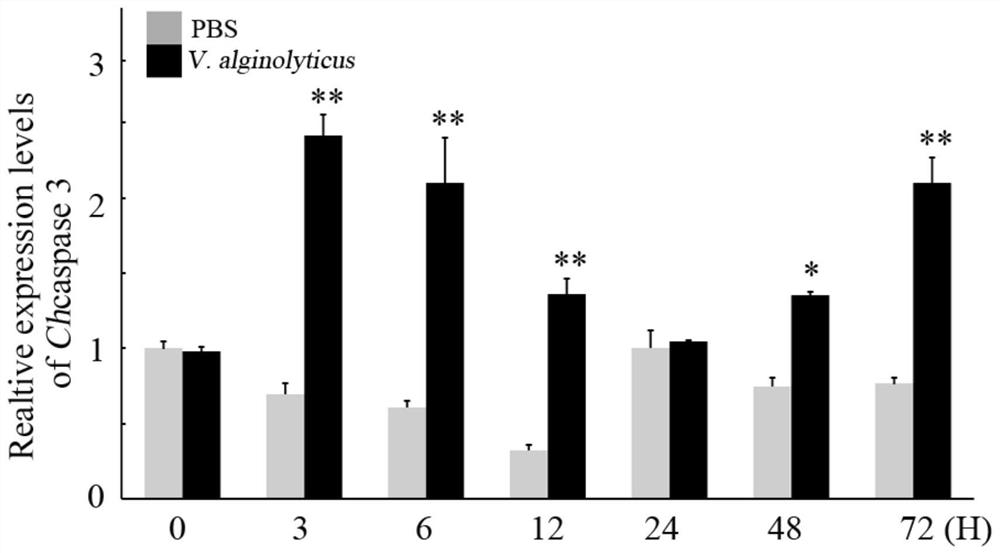
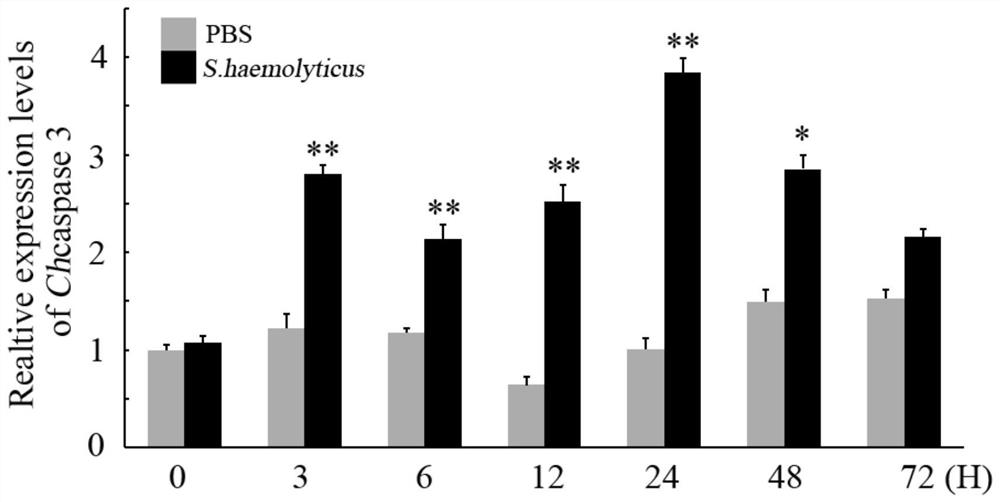
A kind of oyster cell apoptosis gene caspase 3 gene and its application in the preparation of pathological detection diagnostic reagents
A technology for detecting reagents and oysters, which can be used in applications, genetic engineering, plant genetic improvement, etc., and can solve problems such as output fluctuations, high energy consumption, and large breeding areas
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. RNA extraction and reverse transcription
[0032] a). Hong Kong oyster cells or tissues were added with Trizol, ground with a tissue grinder, and left at room temperature for 5 minutes to fully lyse.
[0033] b). Centrifuge at 12,000rpm for 5min and discard the precipitate.
[0034] c). Add chloroform to 200ul chloroform / ml Trizol, vortex and mix well, and place at room temperature for 15min.
[0035] d). Centrifuge at 12,000 g for 15 min at 4°C.
[0036] e). Aspirate the upper aqueous phase into another centrifuge tube.
[0037] f). According to 0.5ml isopropanol / ml Trizol, add isopropanol and mix well, and place at room temperature for 5-10min.
[0038] g). Centrifuge at 12,000g at 4°C for 10 minutes, discard the supernatant, and sink the RNA to the bottom of the tube.
[0039] h). Add 75% ethanol to 1ml 75% ethanol / ml Trizol, shake the centrifuge tube gently, and suspend the precipitate.
[0040] i). Centrifuge at 8,000g at 4°C for 5 minutes, and discard the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com