Isothermal amplification and detection technique based on CRISPR-chain substitution
A technology of isothermal amplification and strand substitution, which is applied in the determination/testing of microorganisms, biochemical equipment and methods, etc., to achieve the effects of improving specificity, rapid response, and rapid recognition response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
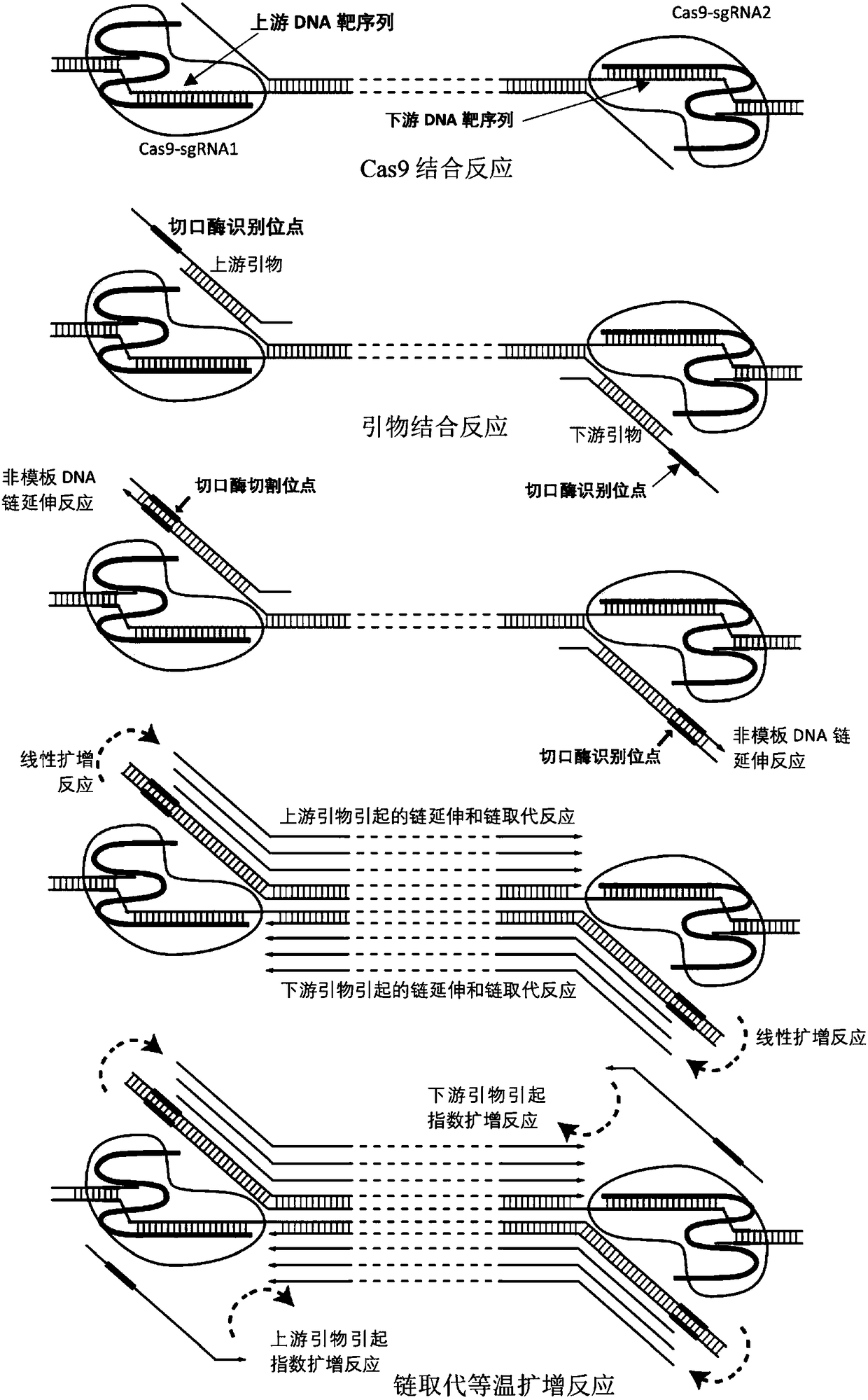
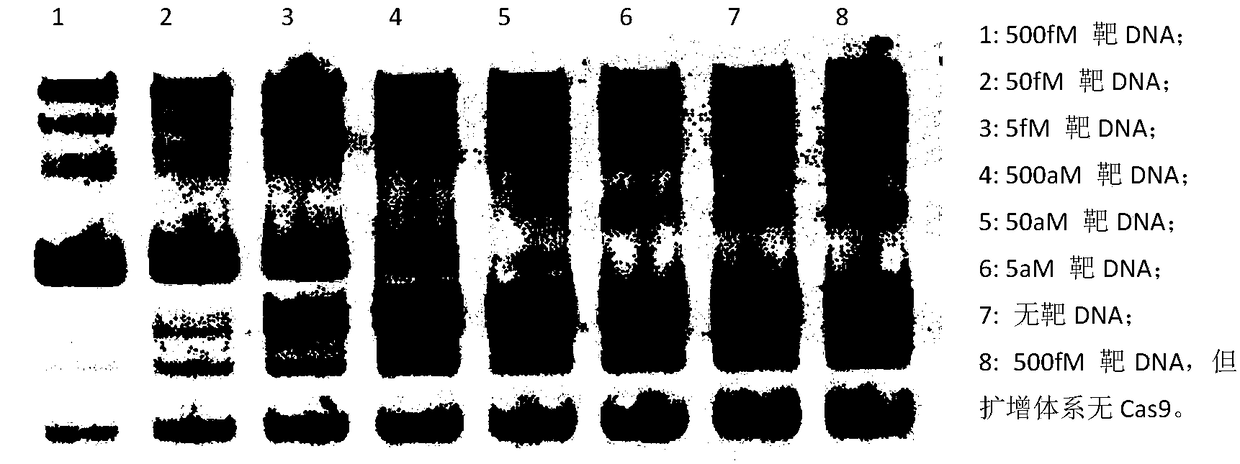
[0069] Example 1. Using the strand replacement amplification reaction based on the CRISPR-Cas9 system to specifically amplify short target DNA double strands, and detect them by traditional PAGE technology
[0070] A pair of sgRNA was designed for a short DNA double-strand (450bp) and synthesized by in vitro transcription, which specifically recognizes the upstream CCAGTGCAAGTGCAGGTGCCAGA (SEQ ID NO:1) of the short DNA double-strand (the 5'end CCA is the complement of the PAM sequence NGG Sequence) and downstream GGCCCAGACTGAGCACGTGATGG (SEQ ID NO: 2) (where the 3'end TGG is the PAM sequence). The sgRNAs that recognize upstream and downstream sequences are named sgRNA-UPS and sgRNA-DNS, respectively.
[0071] sgRNA-UPS: GUGCAAGUGCAGGUGCCAGAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCU (SEQ ID NO: 3)
[0072] sgRNA-DNS: GGCCCAGACUGAGCACGUGAGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCU (SEQ ID NO: 4)
[0073] The strand re...
Embodiment 2
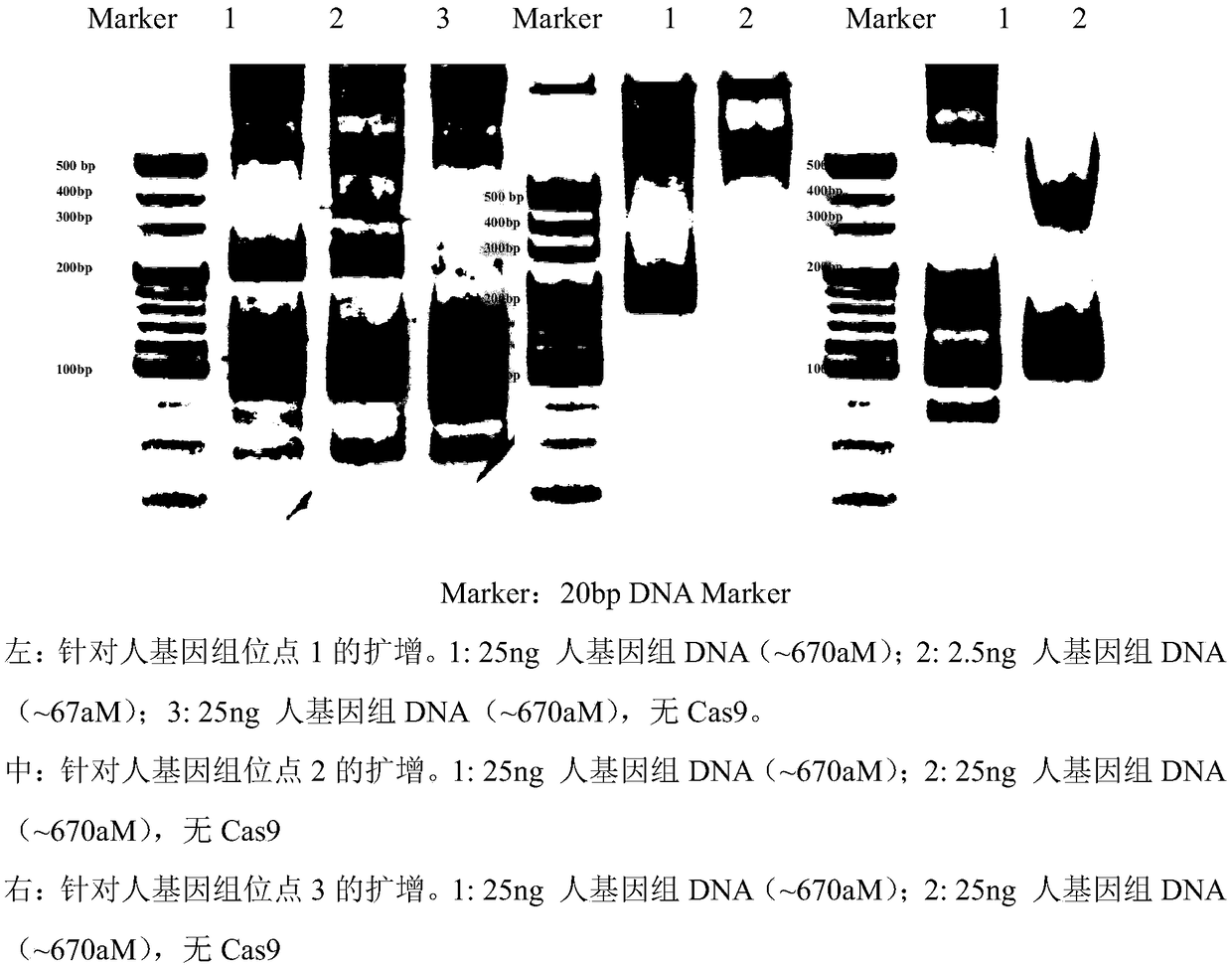
[0084] Example 2. Use the strand replacement amplification reaction based on the CRISPR-Cas9 system to specifically amplify short-term fragments of human genomic DNA, and detect them by traditional PAGE technology
[0085] First, use a commercial kit to extract genomic DNA from human embryonic kidney cells (HEK293). The extracted human genomic DNA was serially diluted to 2.5ng / μL. In the sample to be tested, the concentration of human genomic DNA is approximately 692.4 aM. In the test, a solution sample of plasmid DNA (pEGFP-C2) or mouse genomic DNA was used as a negative control. The concentration of plasmid DNA (pEGFP-C2) or mouse genomic DNA in the negative control was both 30ng / μL.
[0086] Using the same method as in Example 1, for a specific region in the human genome, sgRNA pairs that specifically recognize the upstream and downstream target DNA sequences of the region are designed and synthesized. In order to verify the reproducibility of the chain substitution amplifica...
Embodiment 3
[0111] Example 3. Using the strand replacement amplification reaction based on the CRISPR-Cas9 system to specifically amplify the short target DNA double-strands, and detect them by RT-PCR technology (real-time fluorescent PCR).
[0112] Using the same method as in Example 1, for a short DNA double strand (450 bp), the sgRNA in Example 1 was used to specifically amplify the short DNA double strand sgRNA-UPS and sgRNA-DNS.
[0113] The specific experimental steps are as follows:
[0114] 1. As in the steps 1 to 3 in Example 1, under the same conditions, prepare Cas9-sgRNA protein nucleic acid complexes with targeting activity respectively to complete the upper part of the Cas9-sgRNA protein nucleic acid complex and the short DNA double strands to be detected. Recognition reaction of downstream DNA target sequence and primer binding reaction.
[0115] 2. Add the optimized chain replacement isothermal amplification reaction enzymes in equal volume (final concentration: Klenow Fragmentexo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com