Method for establishing luciferase knock-in cell line based on CRISPR-targeted genome modification technology
A luciferase and cell line technology, applied in the field of molecular biology, can solve problems such as cumbersome process, unfavorable high-throughput screening, and difficulty in accurately reflecting gene expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1: Design, synthesis and vector construction of sgRNA downstream of the stop codon targeting the target gene SREBP1
[0052] (1) Select the downstream of the stop codon of the SREBP1 gene as the targeting region (TSF), with a length of about 1000 bp;
[0053] (2) Find all NGGs and their first 12 bases in the TSF region and perform Blast at NCBI to screen out the sequence that exactly matches the target sequence and the only one (if there is no NGG that meets the requirements, look up CCN in reverse), reducing the potential off-target sites;
[0054] This embodiment designs 4 sgRNAs, wherein:
[0055] The sequence of sgRNA1 is: GTCGAAGCTTTGAAGGCCGA;
[0056] The sequence of sgRNA2 is: GATCTTGACCCTAAGACCGG;
[0057] The sgRNA3 sequence is: GTGGCCGATCGGGGCACTGC;
[0058] The sgRNA4 sequence is: GCTTTCCCGGACTGCAAGCA.
[0059] ACCG was added to its 5' to obtain a forward oligonucleotide, its complementary strand was obtained, and AAAC was added to its 5' to obta...
Embodiment 2
[0060] Example 2: Construction of vectors expressing sgRNA components and Cas9 genes
[0061] (1) After passing the T7E1 detection, select the sgRNA expression vector with high targeting efficiency;
[0062] (2) The sgRNA expression vector with high targeting efficiency and the Cas9 expression vector were digested with KpnI and SpeI respectively, and recovered by agarose gel electrophoresis with a mass concentration of 1%. Excision and sequencing identified positive clones. The obtained clone was named as pCMV-Cas9-SV40pA-U6-sgRNAs-SV40pA (such as figure 1 shown).
Embodiment 3
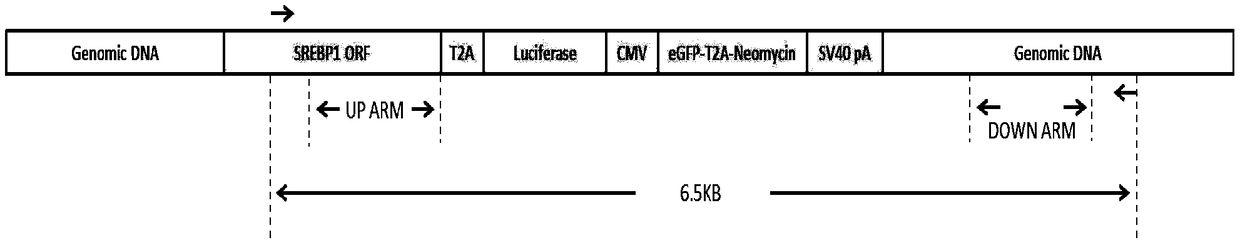
[0063] Example 3: Construction of the targeting vector pUC19 / SREBP1-donor targeting the SREBP1 gene
[0064] The constructed targeting vector contains two sequences of 909 bp upstream and 887 bp downstream of the targeting break site as the upstream and downstream homology arms of the targeting vector, wherein:
[0065] The upstream homology arm was obtained using primers SREBP1up arm ClaI for sequence TATCGATGTCAGGCAGTGGTGGAGATG and SREBP1up arm SpeI reverse sequence GACTAGTGCTGGAAGTGACAGTGGTCC.
[0066] The downstream homology arms were obtained using primers SREBP1down arm SalⅠfor sequence CGTCGACGGCCACAAGGTACACAACTTT and SREBP1down arm BglⅡ reverse reverse sequence AAGATCTCTGTCCGTCCGTGTCCTCA.
[0067] Then connect it to the pU19 expression vector with the T2A-Luciferase reporter gene, the positive screening element CMV-eGFP-T2A-Neomycin-SV40pA and the negative screening element PGK-TK-T2A-mCherry-SV40pA preserved in our laboratory, and the obtained The vector was named pU...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com