Preparation method and applications of humanized gene modified animal models
A humanized, non-human animal technology, applied in the fields of botanical equipment and methods, biochemical equipment and methods, plant genetic improvement, etc., can solve the problems of poor cell survival, short lifespan, and reduced CD4+ T cell proliferation, etc. To save time and cost, speed up the research and development process, and reduce the risk of drug development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0170] Example 1 Homologous Recombination Fragment Primer Design and PCR Amplification
[0171] According to the experimental design scheme, primers for amplifying seven homologous recombination fragments (A1, A2-1, A2-2, A3, B, C1, C2) were designed. The primer sequences are shown in Table 1.
[0172] Table 1 Homologous recombination fragments and their corresponding lengths and primer sequences
[0173]
[0174]
[0175] Using BAC as a template, 7 homologous recombination fragments were amplified using KOD-plus-polymerase. Among them, A1, A3, B, C1, and C2 were selected from Bacterial Artificial Chromosome (BAC) containing the mouse CD137 genome (product number: RPCI23.C, clone ID: 448J14, hereinafter referred to as "bacteria containing mouse BAC") As the template, A2-1 and A2-2 selected the bacterial artificial chromosome containing the human CD137 genome (article number: RPCI11.C, clone ID: 208A7, hereinafter referred to as "bacteria containing human BAC") as the te...
Embodiment 2
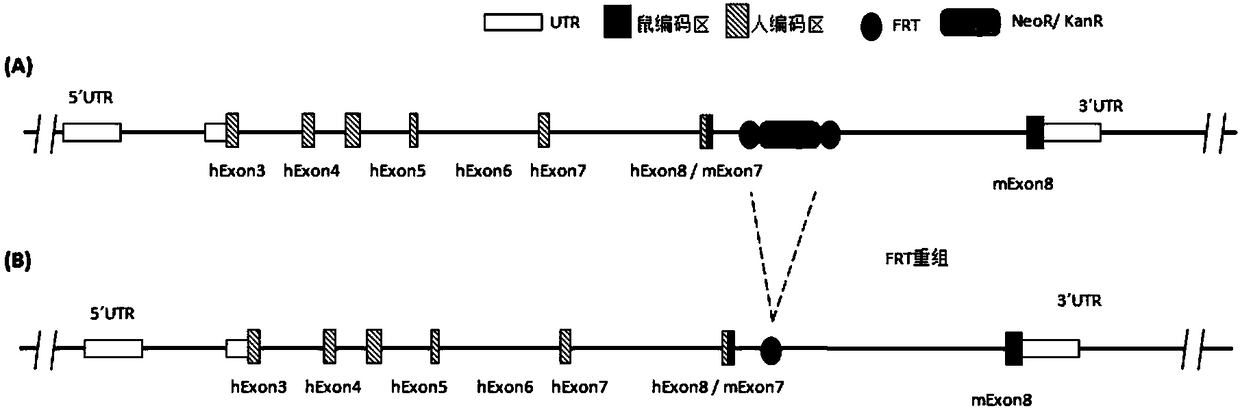
[0179] Example 2 Construction of Homologous Recombination Targeting Vector
[0180] Schematic diagram of comparison between mouse Cd137 gene and human CD137 gene figure 1 shown. The inventor designed figure 2 The targeting strategy shown, figure 2 The design of the targeting vector is also shown. Mouse Cd137 (based on NM_011612.2, ie SEQ ID NO: 15 and its corresponding protein expression NP_035742.1, ie SEQ ID NO: 16) has a total of 8 exons, of which exon 1 does not code, and exon 2 Some are not coded. The present inventors used human CD137 gene (based on NM_001561.5, namely SEQ ID NO: 17 and its corresponding protein expression NP_001552.2, namely SEQ ID NO: 18) for the main part of exon No. 2-7 of mouse Cd137 gene Fragment replacement, and adding neo gene to the targeting vector for positive clone screening. The length of its 5' homology arm (SEQ ID NO:19) is 4847bp, the length of its 3' homology arm is 4728bp (SEQ ID NO:20), and the length of the human DNA fragment ...
Embodiment 3
[0199] Example 3 Verification of carrier pDTA-down-ABC
[0200] Randomly select 4 pDTA-down-ABC clones, and use 3 groups of restriction endonucleases for digestion verification. Among them, 9010bp+5834bp+4165bp+3058bp+1448bp+494bp should appear when using XhoI+NdeI digestion, and using SalI+SmaI Enzyme digestion should produce 11641bp+7563bp+4083bp+722bp, and digestion with BstZ17I+KpnI should produce 12111bp+7897bp+3582bp+419bp. The results of enzyme digestion showed that plasmids 1, 2, and 3 were all in line with expectations, see Figure 5 , Plasmids 1 and 2 were sent to the sequencing company for sequencing and confirmed to be correct.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com