Multi-quenching fluorescent probe and method for detecting target nucleic acid sequence
A technology of target nucleic acid sequence and fluorescent probe, applied in the field of genetic engineering, can solve the problems of superposition of interference, high fluorescence background, affecting the interpretation of dissolution curve, etc., and achieve the effect of clear interpretation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1 Target nucleic acid sequence detection technology based on cleavage of fluorescent quencher group on hybridization sequence
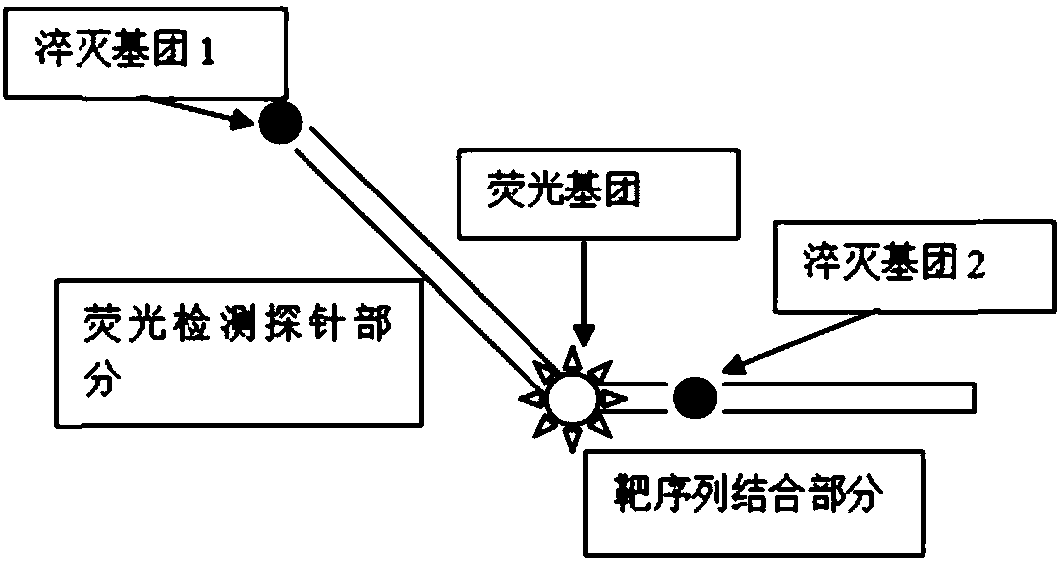
[0020] see figure 1 , is a structural schematic diagram of a multiple quenched fluorescent probe for detecting target nucleic acid sequences of the present invention. The multiple quenched fluorescent probe includes two parts, and the 5' end is a fluorescent detection probe part, which includes a quenching group 1 and a fluorescent group This part is not combined with the target target sequence to be detected, and the 3' end is the target sequence binding part, which contains other quenching groups other than quencher group 1, and this part is reverse complementary to the target target sequence to be detected combined.
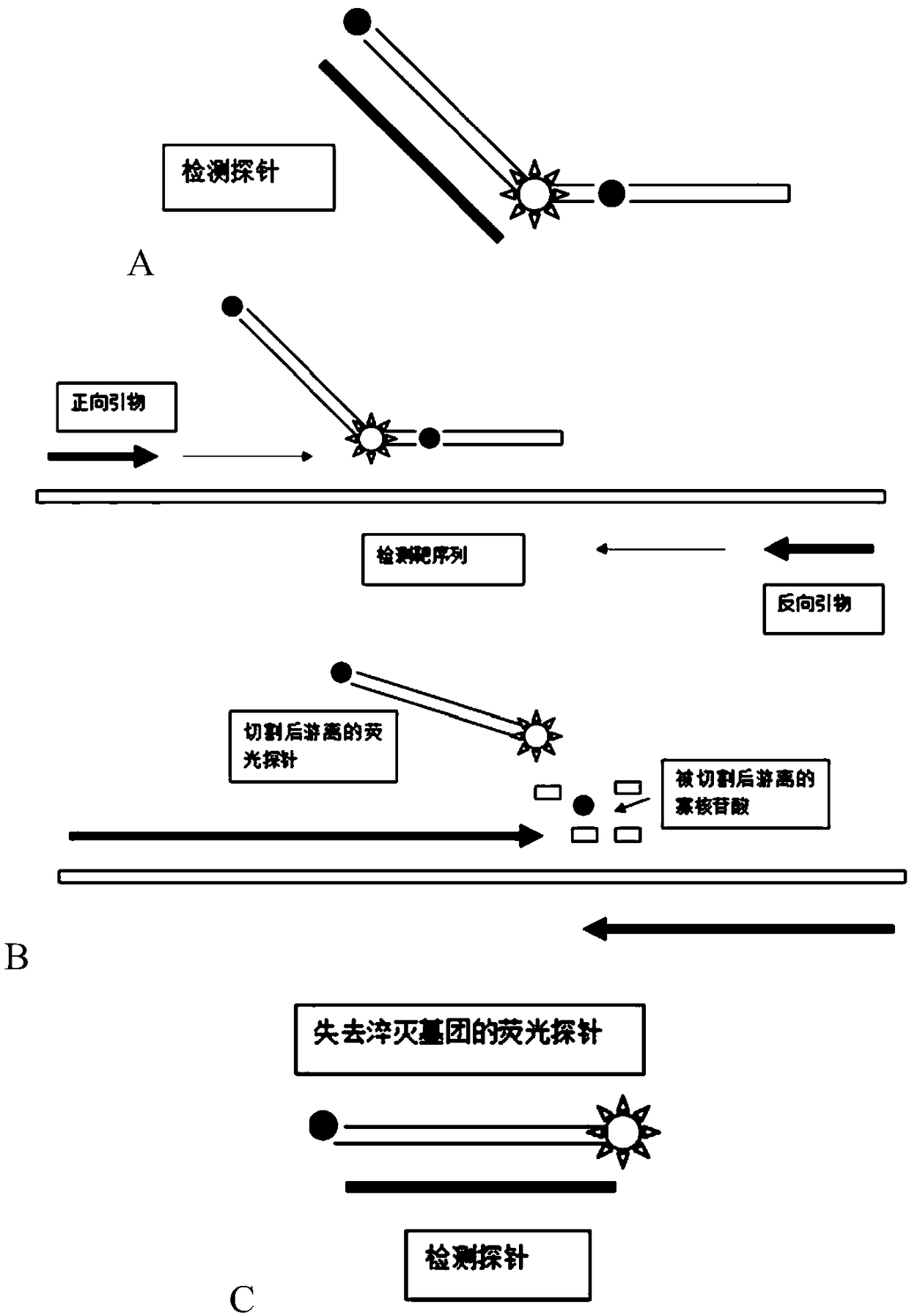
[0021] see figure 2 A-C, are schematic diagrams of the target nucleic acid sequence detection method based on the cleavage of the hybridization sequence fluorescence quencher group of the present invention. When the...
Embodiment 2
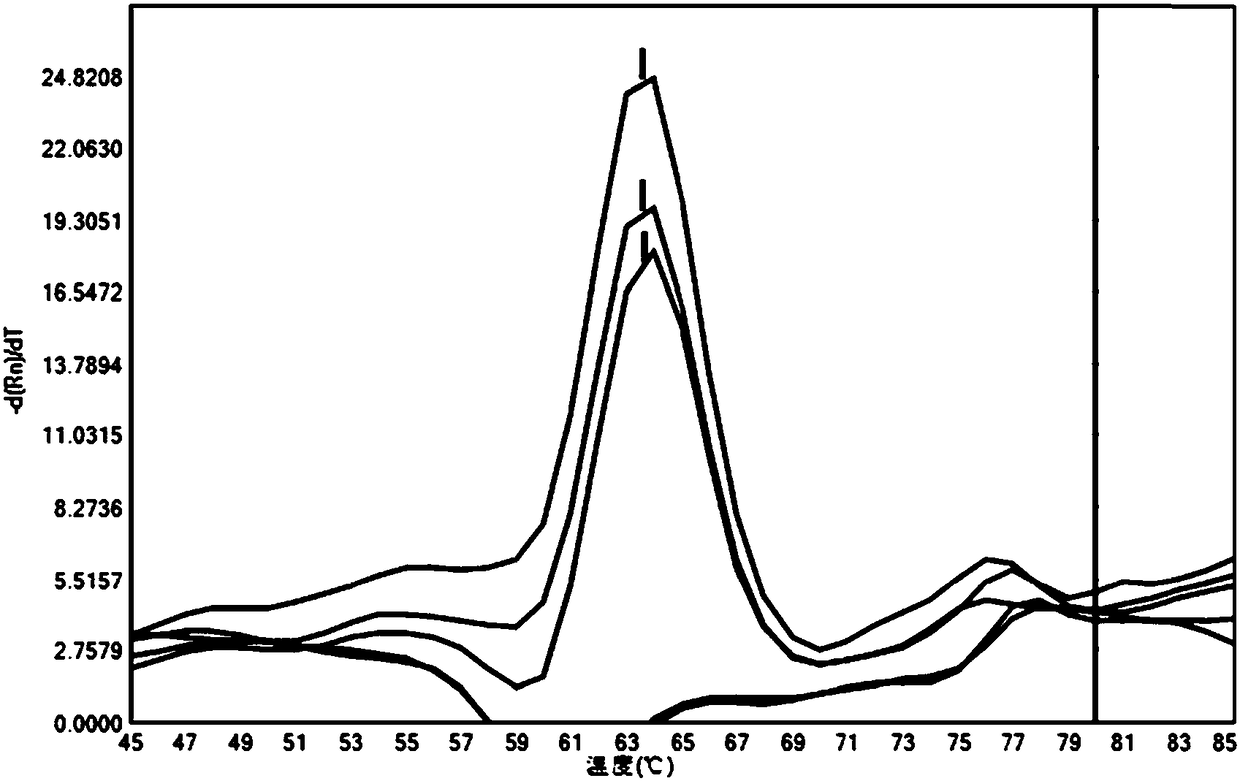
[0022] Example 2 Using the method of Example 1 of the present invention to detect different concentrations of target sequence templates
[0023] The method of Example 1 of the present invention was used to detect different concentrations of target sequence templates to evaluate its practicability. The target sequence template is an artificial plasmid (T vector plasmid, synthesized by Sangon Biotech (Shanghai) Co., Ltd.) containing the PCR amplification region, which is diluted to different concentrations of E2, E3, and E4 according to the concentration (10 2 copies / ul, 10 3 copies / ul, 10 4 copies / ul).
[0024] The instrument used is Shanghai Hongshi Slan-96p fluorescence quantitative PCR instrument.
[0025] The amplification primer sequences are:
[0026] NG-F:TACGCCTGCTACTTTCACGCT (SEQ ID No:1)
[0027] NG-R:CAATGGATCGGTATCACTCGC (SEQ ID No:2)
[0028] The fluorescent probe sequence is:
[0029] NG-P:ACGACGGCTTGGCTTTACTTCATGCCCCTCATTGGCGTGTTTCG
[0030] (SEQ ID No: 3...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com